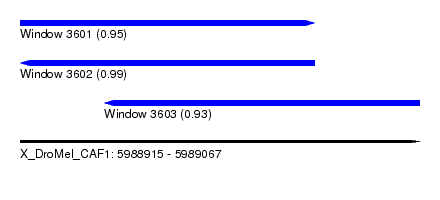
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,988,915 – 5,989,067 |
| Length | 152 |
| Max. P | 0.992355 |

| Location | 5,988,915 – 5,989,027 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -23.69 |
| Energy contribution | -24.92 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5988915 112 + 22224390 AAGUUGAACUUAAAUUAGUUUAGCCAAAGCUGUUUGAUUGCUUCUCGUACCGAACUGAAAUGCAACCAGGGCCAAGUUGCACCGCAGCCCAGCCACAAAAUCACUUUGGCUC ....((((((......))))))(((((((...((((.((((...(((........)))...))))....(((...(((((...)))))...))).))))....))))))).. ( -28.60) >DroSec_CAF1 4033 112 + 1 AAGUUGAACUCAAAUUAGUUUAGCCAAAGCUGUUUGAUUGCUUCUCGUACCGAACUAAAAUGCAACCAGGGCAAAGUUGCACCGCAGCCCAGCCACAAAAUCACUUUGGCUC ....((((((......))))))((((((((((.((((((.....(((...))).....))).))).)))(((...(((((...)))))...))).........))))))).. ( -26.80) >DroSim_CAF1 5655 112 + 1 AAGUUGAACUUAAAUUAGUUUAGCCAAAGCUGUUUGAUUGCUUCUCGUACCGAACUGAAAUGCAACCAGGGCAAAGUUGCACCGCAGCCCAGCCACAAAAUCACUUUGGCUC ....((((((......))))))(((((((...((((.((((...(((........)))...))))....(((...(((((...)))))...))).))))....))))))).. ( -28.60) >DroEre_CAF1 3887 90 + 1 AAGUUGCACUUAAAUUAGUUUAGCCAAAGCAGUUUGAUU----------------------GCAACCAGGGCGAAGUUGCACCGCAGCCCAGCCACAAAAUCGCUGCGGCUG ..((((((.(((((((.((((.....))))))))))).)----------------------)))))..(((((....))).)).(((((((((.........)))).))))) ( -30.20) >DroYak_CAF1 6018 112 + 1 AAGUUGAACUUAAAUUAGUUUAGCCAAAGCAGUUUGAUUGCUUCUCAUACCAAAUUGAAAUGCAACCAGGGCGAAGUUGCACCGCAGCCCAGCCACAAAAUCACUUUGGCUC ....((((((......))))))(((((((...((((.((((...(((........)))...))))....(((...(((((...)))))...))).))))....))))))).. ( -31.20) >consensus AAGUUGAACUUAAAUUAGUUUAGCCAAAGCUGUUUGAUUGCUUCUCGUACCGAACUGAAAUGCAACCAGGGCAAAGUUGCACCGCAGCCCAGCCACAAAAUCACUUUGGCUC ....((((((......))))))(((((((...((((.((((...(((........)))...))))....(((...(((((...)))))...))).))))....))))))).. (-23.69 = -24.92 + 1.23)


| Location | 5,988,915 – 5,989,027 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5988915 112 - 22224390 GAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUGGCCCUGGUUGCAUUUCAGUUCGGUACGAGAAGCAAUCAAACAGCUUUGGCUAAACUAAUUUAAGUUCAACUU .(((((((((..(((((((.((((((((..((((((((.......))))))))..))))))))))))))).(........).))))))))).((((......))))...... ( -39.70) >DroSec_CAF1 4033 112 - 1 GAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUUGCCCUGGUUGCAUUUUAGUUCGGUACGAGAAGCAAUCAAACAGCUUUGGCUAAACUAAUUUGAGUUCAACUU .(((((((((..(((((((.((((((((..((((((((.......))))))))..))))))))))))))).(........).))))))))).((((......))))...... ( -38.10) >DroSim_CAF1 5655 112 - 1 GAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUUGCCCUGGUUGCAUUUCAGUUCGGUACGAGAAGCAAUCAAACAGCUUUGGCUAAACUAAUUUAAGUUCAACUU .(((((((((..(((((((.((((((((..((((((((.......))))))))..))))))))))))))).(........).))))))))).((((......))))...... ( -39.70) >DroEre_CAF1 3887 90 - 1 CAGCCGCAGCGAUUUUGUGGCUGGGCUGCGGUGCAACUUCGCCCUGGUUGC----------------------AAUCAAACUGCUUUGGCUAAACUAAUUUAAGUGCAACUU .(((((.((((...(((..((..((..((((.......))))))..))..)----------------------))......)))).)))))..................... ( -27.90) >DroYak_CAF1 6018 112 - 1 GAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUCGCCCUGGUUGCAUUUCAAUUUGGUAUGAGAAGCAAUCAAACUGCUUUGGCUAAACUAAUUUAAGUUCAACUU .(((((((((((...((..((..((..((((.......))))))..))..))..)).....(((.(((.......))).)))))))))))).((((......))))...... ( -34.10) >consensus GAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUCGCCCUGGUUGCAUUUCAGUUCGGUACGAGAAGCAAUCAAACAGCUUUGGCUAAACUAAUUUAAGUUCAACUU .(((((((((........(((..((.(((...))).))..))).(((((((.((((.((.....)))))).)))))))....)))))))))..................... (-29.40 = -29.62 + 0.22)

| Location | 5,988,947 – 5,989,067 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -35.29 |
| Energy contribution | -35.85 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5988947 120 - 22224390 AAAGUGUGGUUGAGUCGAGUCGAGUUGAACUGUAAGCUGGGAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUGGCCCUGGUUGCAUUUCAGUUCGGUACGAGAAGCAAUCAAA ......((((((..(((..((((..((((.(((((.(..((.(((((..((...(((..((...))..)))..))..)))))))..)))))).))))..))))..)))....)))))).. ( -45.20) >DroSec_CAF1 4065 120 - 1 AAAGUGUGGUUGAGUCGAGUCGAGUUGAACUGUAAGCUGGGAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUUGCCCUGGUUGCAUUUUAGUUCGGUACGAGAAGCAAUCAAA ......((((((..(((..((((..((((.(((((.(..((.((.(((((....(((..((...))..)))....)))))))))..)))))).))))..))))..)))....)))))).. ( -39.70) >DroSim_CAF1 5687 120 - 1 AAAGUGUGGUCGAGUCGAGUCGAGUUGAACUGUAAGCUGGGAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUUGCCCUGGUUGCAUUUCAGUUCGGUACGAGAAGCAAUCAAA ....((..(((((.(((...))).))).))..)).(((((...)).......(((((((.((((((((..((((((((.......))))))))..))))))))))))))))))....... ( -38.50) >DroYak_CAF1 6050 116 - 1 AAAGUGUGGUUG----GAAUUGAGUGGAAGUGUAAGCUGGGAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUCGCCCUGGUUGCAUUUCAAUUUGGUAUGAGAAGCAAUCAAA ....(((..((.----(.((..(((.(((((((((.(..((.((..((((....(((..((...))..)))....)))).))))..)))))))))).)))..)).).))..)))...... ( -34.00) >consensus AAAGUGUGGUUGAGUCGAGUCGAGUUGAACUGUAAGCUGGGAGCCAAAGUGAUUUUGUGGCUGGGCUGCGGUGCAACUUUGCCCUGGUUGCAUUUCAGUUCGGUACGAGAAGCAAUCAAA ......((((((..(((..((((((((((.(((((.(..((.((.(((((....(((..((...))..)))....)))))))))..)))))).))))))))))..)))....)))))).. (-35.29 = -35.85 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:08 2006