
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,987,894 – 5,988,024 |
| Length | 130 |
| Max. P | 0.938275 |

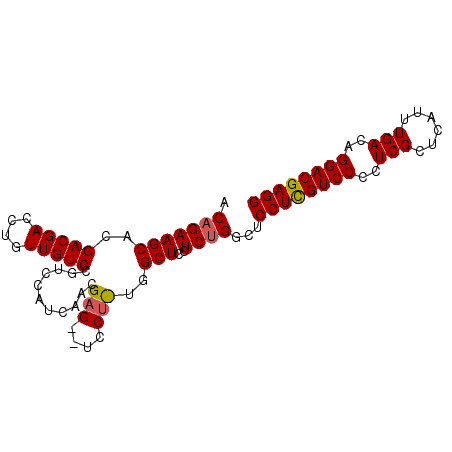
| Location | 5,987,894 – 5,987,984 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5987894 90 + 22224390 ACAGAAGCACGACGACCUGUUGUCCGUCCAUCAACGAGCUUCCUUUGGCUCCUC-GUCCCCUUGUCCAUUCGACCAUUUGACAGGACGAGG ..........(((((...((((.........))))(((((......))))).))-))).((((((((..((((....))))..)))))))) ( -28.20) >DroSec_CAF1 3075 72 + 1 ACAGAAGCACGACGACCUGUUGUCCG----------A---------GGCUCCUCUGGCUCCUCGUCCCCUCGCUCAUUUGACAGGACGAGG .(((((((..(((((....)))))..----------.---------.)))..))))...((((((((..(((......)))..)))))))) ( -26.60) >DroSim_CAF1 4628 91 + 1 ACAGAAGCACGACGACCUGUUGUCCGUCCAUCAACGAGGCUCCUCUGGCUCCUCUGGCUCCUCGUCCCCUCGCUCAUUUGACAGGACGAGG .(((((((..((((..(....)..)))).......(((....)))..)))..))))...((((((((..(((......)))..)))))))) ( -30.20) >consensus ACAGAAGCACGACGACCUGUUGUCCGUCCAUCAACGAG__UCCU_UGGCUCCUCUGGCUCCUCGUCCCCUCGCUCAUUUGACAGGACGAGG .(((((((..(((((....)))))...........(((....)))..)))..))))...((((((((..(((......)))..)))))))) (-22.26 = -22.77 + 0.50)

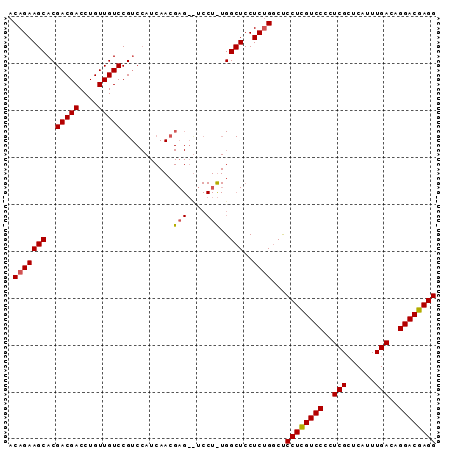
| Location | 5,987,894 – 5,987,984 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -24.90 |
| Energy contribution | -27.07 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5987894 90 - 22224390 CCUCGUCCUGUCAAAUGGUCGAAUGGACAAGGGGAC-GAGGAGCCAAAGGAAGCUCGUUGAUGGACGGACAACAGGUCGUCGUGCUUCUGU (((((((((.(......(((.....))).).)))))-)))).......((((((....(((((..(........)..))))).)))))).. ( -31.50) >DroSec_CAF1 3075 72 - 1 CCUCGUCCUGUCAAAUGAGCGAGGGGACGAGGAGCCAGAGGAGCC---------U----------CGGACAACAGGUCGUCGUGCUUCUGU (((((((((.((........)).)))))))))...((((((.((.---------.----------((..(....)..))..)).)))))). ( -31.50) >DroSim_CAF1 4628 91 - 1 CCUCGUCCUGUCAAAUGAGCGAGGGGACGAGGAGCCAGAGGAGCCAGAGGAGCCUCGUUGAUGGACGGACAACAGGUCGUCGUGCUUCUGU (((((((((.((........)).)))))))))...((((((.((..(((....))).......((((..(....)..)))))).)))))). ( -37.90) >consensus CCUCGUCCUGUCAAAUGAGCGAGGGGACGAGGAGCCAGAGGAGCCA_AGGA__CUCGUUGAUGGACGGACAACAGGUCGUCGUGCUUCUGU (((((((((.((........)).)))))))))......(((((((.(((....))).......((((..(....)..))))).)))))).. (-24.90 = -27.07 + 2.17)

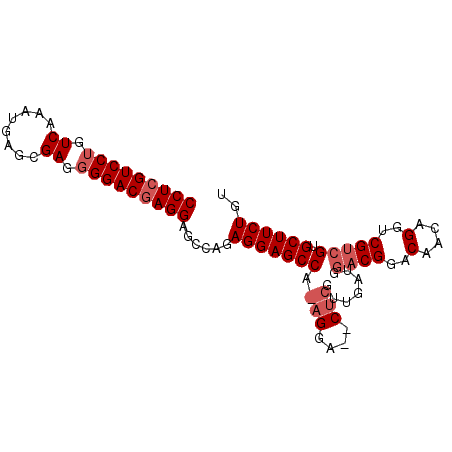

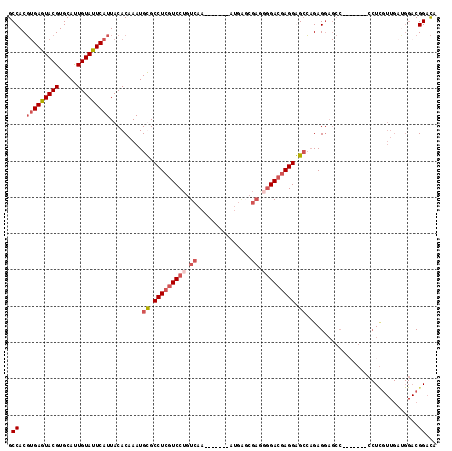
| Location | 5,987,914 – 5,988,024 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.66 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -21.37 |
| Energy contribution | -23.43 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5987914 110 - 22224390 GCCACGUGAAUACGUGCAUUGUAUUCAUUACACAAAUGCGCCUCGUCCUGUCAA-------AUGGUCGAAUGGACAAGGGGAC-GAGGAGCCAAAGGAAGCUCGUUGAUGGACGGACA .((..(((((((((.....))))))))).........((.(((((((((.(...-------...(((.....))).).)))))-)))).))....))..(..((((....))))..). ( -35.30) >DroSec_CAF1 3095 92 - 1 GCCACGUGAGUACGUGCAUUGUAUUCAUUACACAAAUGUGCCUCGUCCUGUCAA-------AUGAGCGAGGGGACGAGGAGCCAGAGGAGCC---------U----------CGGACA .....(((((((((.....)))))))))..(((....)))(((((((((.((..-------......)).))))))))).(((.(((....)---------)----------))).). ( -35.00) >DroSim_CAF1 4648 111 - 1 GCCACGUGAGUACGUGCAUUGUAUUCAUUACACAAAUGUGCCUCGUCCUGUCAA-------AUGAGCGAGGGGACGAGGAGCCAGAGGAGCCAGAGGAGCCUCGUUGAUGGACGGACA .((..(((((((((.....)))))))))..(((((..((.(((((((((.((..-------......)).))))))))).))..((((..((...))..)))).))).))...))... ( -41.10) >DroYak_CAF1 4999 98 - 1 GCCACU-GAGUACGUGCACUGUAUUCAUUACACAAAUGCGCCUCGUCCCGUCAGGAAGGAGAGC--------GAGGAGGGGCGAGAG-----------UCCUUGUUGUGGGACGGAUA .((..(-(((((((.....))))))))...(((((.....(((((((((........)).)).)--------))))((((((....)-----------))))).)))))....))... ( -34.20) >consensus GCCACGUGAGUACGUGCAUUGUAUUCAUUACACAAAUGCGCCUCGUCCUGUCAA_______AUGAGCGAGGGGACGAGGAGCCAGAGGAGCC_______CCUCGUUGAUGGACGGACA .((..(((((((((.....))))))))).........((.(((((((((.((...............)).))))))))).))...............................))... (-21.37 = -23.43 + 2.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:05 2006