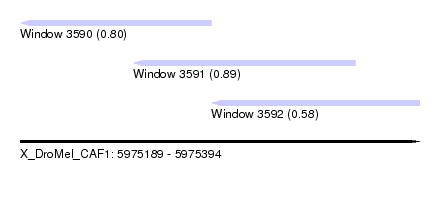
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,975,189 – 5,975,394 |
| Length | 205 |
| Max. P | 0.893205 |

| Location | 5,975,189 – 5,975,287 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.94 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5975189 98 - 22224390 GAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAGGAUUUGUGCUCAGGUGUCCUUGGACUAAUUAAAAGCCUUUAUGG---CUCACAGACACUCA------------------- (((((........(((((..(((........)))..))))).((((((.......(((....)))........((((.....))---))))))))))))).------------------- ( -33.50) >DroPse_CAF1 5674 101 - 1 GAGUGGACUCCAGUCUGUUGUUGUGUCAUUCGGAUGGCAGGAUUUGUGCUCAGGUGUCCUUGGGCCAAUUAAAAGCCUUUAAGUGCAUACACAGACACAGC------------------- (((....)))..((((((.((.(((.((((..((.(((.....(((.(((((((....))))))))))......)))))..))))))))))))))).....------------------- ( -32.80) >DroSim_CAF1 9223 98 - 1 GAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAGGAUUUGUGCUCAGGUGUCCUUGGACUAAUUAAAAGCCUUUAUGG---CUCACAGACACUCA------------------- (((((........(((((..(((........)))..))))).((((((.......(((....)))........((((.....))---))))))))))))).------------------- ( -33.50) >DroYak_CAF1 9274 98 - 1 GAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAGGAUUUGUGCUCAGGUGUCCUUGGACUAAUUAAAAGCCUUUAUGG---CUCACAGACACUCA------------------- (((((........(((((..(((........)))..))))).((((((.......(((....)))........((((.....))---))))))))))))).------------------- ( -33.50) >DroPer_CAF1 9258 120 - 1 GAGUGGACUCCAGUCUGUUGUUGUGUCAUUCGGAUGGCAGGAUUUGUGCUCAGGUGUCCUUGGGCCAAUUAAAAGCCUUUAAGUGCAUACACAGACACAGCGUACAGACGUACAGACUGA (((....)))((((((...((((((((........(((.....(((.(((((((....))))))))))......))).....(((....))).))))))))((((....)))))))))). ( -42.70) >consensus GAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAGGAUUUGUGCUCAGGUGUCCUUGGACUAAUUAAAAGCCUUUAUGG___CUCACAGACACUCA___________________ (((((........(((((..(((........)))..))))).((((((...(((((((....))).........))))...........))))))))))).................... (-21.46 = -21.94 + 0.48)

| Location | 5,975,247 – 5,975,361 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -28.32 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5975247 114 - 22224390 ----ACAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCACCCGGCGACUGUCCACUUUUGCCAGUGCCAUCUUCGAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAG ----....................((((((........((.(((..(((((..(....)..))))).))).)).....))))))........((((..(((........)))..)))) ( -27.62) >DroSim_CAF1 9281 114 - 1 ----CCAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCACCCGGCGACUGUCCACUUUCGCCAGUGCCAUCUUCGAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAG ----....................((((((........((.(((..(((((..(....)..))))).))).)).....))))))........((((..(((........)))..)))) ( -29.72) >DroYak_CAF1 9332 118 - 1 UUUUCCAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCAACCGGCGACUGUCCACUUUCGCCAGUGCCAUCUUCGAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAG ..........(((((((.....((.((((..((.....))......(((((..(....)..))))))))).)).....))))))).......((((..(((........)))..)))) ( -28.80) >consensus ____CCAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCACCCGGCGACUGUCCACUUUCGCCAGUGCCAUCUUCGAGUGGACUCCAGUCUGUUGUGCUGUCAUUUGCAUGGCAG ..........(((((((.....((.((((..((.....))......(((((..(....)..))))))))).)).....))))))).......((((..(((........)))..)))) (-28.32 = -28.10 + -0.22)


| Location | 5,975,287 – 5,975,394 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.88 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -16.02 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5975287 107 - 22224390 CAGGCACCCCGAGCCUCGAAU----C-CCUUC--CCGCCG----ACAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCACCCGGCGACUGUCCACUUUUGCCAGUGCCAUCUUC ..(((((..((.((...(((.----.-..)))--..))))----..........................................(((((..(....)..))))).)))))...... ( -18.50) >DroSim_CAF1 9321 107 - 1 CAGGCACCCCGAGCCCCGAGU----C-CCUUC--CCGCCG----CCAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCACCCGGCGACUGUCCACUUUCGCCAGUGCCAUCUUC ..(((((...(.((..((.(.----.-....)--.))..)----))........................................(((((..(....)..))))).)))))...... ( -20.00) >DroYak_CAF1 9372 118 - 1 CAGGCACCCUGAACCUGUUCUCUUUCCUCUUCCUCCGCCGUUUUCCAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCAACCGGCGACUGUCCACUUUCGCCAGUGCCAUCUUC ..(((((...(((....)))..................................................................(((((..(....)..))))).)))))...... ( -18.60) >consensus CAGGCACCCCGAGCCUCGAAU____C_CCUUC__CCGCCG____CCAUUUUCCAUUUCCAUUUGCCACUUUCACAUUUUGCCACCCGGCGACUGUCCACUUUCGCCAGUGCCAUCUUC ..(((((.............................................((........))......................(((((..(....)..))))).)))))...... (-16.02 = -15.80 + -0.22)

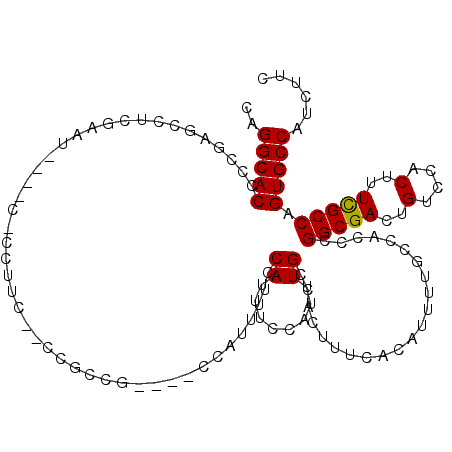
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:58 2006