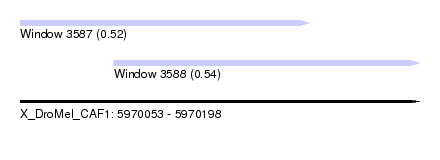
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,970,053 – 5,970,198 |
| Length | 145 |
| Max. P | 0.544361 |

| Location | 5,970,053 – 5,970,158 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -26.05 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5970053 105 + 22224390 UCAGUGGUUCCCCCAGU------UCCAGCUCCUCCACUUGCACCGCCAUUUUGGUGGUUGUUAUUGCCACUGCCGGAUCCUGU------CGCUGCU---GCAGCUGCUGCGGCGGCCGCC ...(((((.....(((.------(((.((.....((...((((((((.....))))).)))...)).....)).)))..))).------((((((.---((....)).))))))))))). ( -38.00) >DroSim_CAF1 4164 114 + 1 UCAGUGGUUCCCCCAGU------CCCAGCUCCUCCAAUUGCACCGCCAUUUUGGUGGUUGUUAUUGCCACCGGCGGAUCCUGUAGCCGCCGCUGCUGCAGCAGCUGCUGCGGCGGCCGCC ...((((((....(((.------....((..........)).(((((.....((((((.......)))))))))))...))).)))))).(((((((((((....))))))))))).... ( -49.00) >DroYak_CAF1 4168 108 + 1 UCAUUGGUUCCCCCAGUUCCAGUUCCAGCUCCGCCACUUGCACCGCCGUUCUGGUGACUGUUAUUGCUACCGCCGGAUCCGGUUGCCG------CU------GCUGCUGCGGCCGCCACC .....((.(((..(.((..((((..(((...(((((...((......))..))))).)))..))))..)).)..))).))(((.((((------(.------......))))).)))... ( -35.00) >consensus UCAGUGGUUCCCCCAGU______UCCAGCUCCUCCACUUGCACCGCCAUUUUGGUGGUUGUUAUUGCCACCGCCGGAUCCUGU_GCCG_CGCUGCU___GCAGCUGCUGCGGCGGCCGCC ...(((((..((.((((.......((.((..........))...........((((((.......))))))...))..............(((((....))))).)))).))..))))). (-26.05 = -27.00 + 0.95)

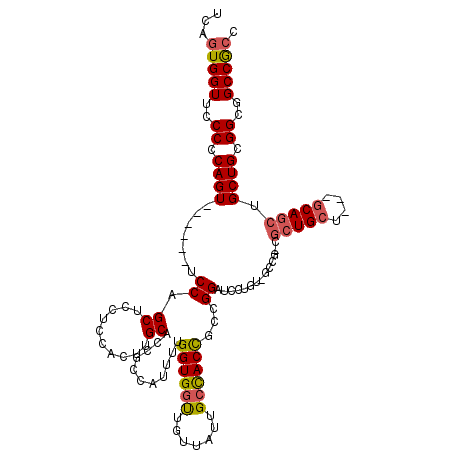

| Location | 5,970,087 – 5,970,198 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -52.37 |
| Consensus MFE | -34.21 |
| Energy contribution | -34.83 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5970087 111 + 22224390 CACCGCCAUUUUGGUGGUUGUUAUUGCCACUGCCGGAUCCUGU------CGCUGCU---GCAGCUGCUGCGGCGGCCGCCGUGCCGGCAACUGUUGCGACUGUUGGAGCCACCGUUGUGG .....((((..((((((((.(..((((.((((((((...(.((------.((((((---((((...)))))))))).)).)..))))))...)).)))).....).))))))))..)))) ( -51.10) >DroSim_CAF1 4198 120 + 1 CACCGCCAUUUUGGUGGUUGUUAUUGCCACCGGCGGAUCCUGUAGCCGCCGCUGCUGCAGCAGCUGCUGCGGCGGCCGCCGUGCCGGCAACUGUUGCAACUGUUGGAGCUACCGUAGUGG ..(((((.....((((((.......))))))))))).........((((..(.(((.((((((.(((.((((..((((......))))..)))).))).)))))).)))....)..)))) ( -55.30) >DroYak_CAF1 4208 108 + 1 CACCGCCGUUCUGGUGACUGUUAUUGCUACCGCCGGAUCCGGUUGCCG------CU------GCUGCUGCGGCCGCCACCGUGCCAGCAACUGUGGCAACUGUGGGGGCCGCCGUUGUGG ....((((.(((((((...((....))...)))))))..))))..(((------(.------...((.((((((.((((..(((((.(....))))))...)))).)))))).)).)))) ( -50.70) >consensus CACCGCCAUUUUGGUGGUUGUUAUUGCCACCGCCGGAUCCUGU_GCCG_CGCUGCU___GCAGCUGCUGCGGCGGCCGCCGUGCCGGCAACUGUUGCAACUGUUGGAGCCACCGUUGUGG .....((((..((((((((((....))..((..(((..............(((((....)))))(((..(((..((((......))))..)))..))).)))..))))))))))..)))) (-34.21 = -34.83 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:55 2006