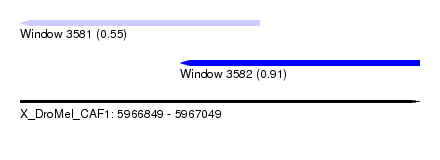
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,966,849 – 5,967,049 |
| Length | 200 |
| Max. P | 0.910339 |

| Location | 5,966,849 – 5,966,969 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -15.60 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5966849 120 - 22224390 UAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUAAAUAAGUGUGUAUCCUAGCUUCUUUUAGCGUUAACCGACACUGCAACACACACUUAUACUCGUACACACUACCAAACACU ..(.((((.(((.......)))...................((((((((((......(((......)))(((....)))........))))))))))..........)))).)....... ( -17.60) >DroSim_CAF1 942 113 - 1 UAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUAAAUAAGUGUGUAUCCUAGCUUCUUUUAGCGUUAACCGACACUGCAACACACAUUUAU------ACACACUACCAAA-ACU ..(.((((.(((.......)))...................((((((((((......(((......)))(((....)))........))))))))))------....)))).)...-... ( -15.30) >DroYak_CAF1 911 117 - 1 UAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUAAAUAAGUGUGUAUCCUAGCUUCUUUUAGCGUUAACCAACAAUGCAACACACAU--AUACU-ACACACACUCCCAAACACA ..(((.((((((.....((((((((((..((.....))..)))))).))))......(((......))))))))).)))..............--.....-................... ( -13.90) >consensus UAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUAAAUAAGUGUGUAUCCUAGCUUCUUUUAGCGUUAACCGACACUGCAACACACAUUUAUACU___ACACACUACCAAACACU ..(((.((((((.....((((((((((..((.....))..)))))).))))......(((......))))))))).)))......................................... (-14.39 = -14.17 + -0.22)


| Location | 5,966,929 – 5,967,049 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -19.92 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5966929 120 - 22224390 AACAUCCAAUCAUGUGAAGCUCUUUUGGUUAAACACACUACAACUACAGCAAUAACAGCCACAGAAACAAUUAGUUAGCUUAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUA .............(((((((..(((((((((.....((((..((((.(((..((((.................))))))))))))))))))))))))..))))))).............. ( -21.03) >DroSim_CAF1 1015 120 - 1 AACAUCCAAUCAUUUGAAGCUCUUUUGGUUAAACACACUACAACUACAGCAAUAACAGCCACAGAAACAAUUAGUUAGCUUAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUA ..............((((((..(((((((((.....((((..((((.(((..((((.................))))))))))))))))))))))))..))))))............... ( -20.13) >DroYak_CAF1 988 120 - 1 GACAUCCAAUCAUUUGAAGCUCUUUUGGUUAAAUACACUACAAUUACAGCAAUAACAGCCACAGAAACAAUUAGUUCGCUUAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUA ..............((((((..((((((((((....((((........((.......))....(((........)))...))))...))))))))))..))))))............... ( -18.60) >consensus AACAUCCAAUCAUUUGAAGCUCUUUUGGUUAAACACACUACAACUACAGCAAUAACAGCCACAGAAACAAUUAGUUAGCUUAGUUAGUUAGCCAAAAACGCUUUAUUCCAUUUCAAAUUA ..............((((((..(((((((((.....((((..((((.(((..((((.................))))))))))))))))))))))))..))))))............... (-18.82 = -18.93 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:49 2006