
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,960,646 – 5,960,806 |
| Length | 160 |
| Max. P | 0.996881 |

| Location | 5,960,646 – 5,960,766 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5960646 120 + 22224390 CUCUACCUCUGAACUGAAUCCCUCAACCAAAAUGGCCACCAAUAAAUUAAAUAGCACAUAAUUGCCGAACGUCAUUAGUGCCACAAAGUACAAUGCGGCCCAAUGACUUGUCUUUUCCAU ...............................((((..................(((......))).((..((((((.(.(((.((........)).)))).))))))...))....)))) ( -16.50) >DroGri_CAF1 19170 120 + 1 CUCUACCUCUGAACUGAAUCCCUCAACCAAAAUGGCCACCAAUAAAUUAAACAGCACAUAAUUGCCGAACGUCAUUAGGGCCACAAAGUACAGUGCGGCCCAAUGACUUGUCUUUUCCAU ...............................((((..................(((......))).((..((((((.(((((.((........)).)))))))))))...))....)))) ( -23.00) >DroWil_CAF1 8749 120 + 1 CUCUACCUCUGAACUGAAUCCCUCAACCAAAAUGGCCACCAAUAAAUUAAAUAGCACAUAAUUGCCGAAUGUCAUUAGGGCCACAAAAUACAAUGCGGCCCAAUGACUUGUCUUUUCCAU ...............................((((..................(((......))).((..((((((.(((((.((........)).)))))))))))...))....)))) ( -23.70) >DroMoj_CAF1 22381 120 + 1 CUCUACCUCUGAACUGAAUCCCUCAACCAAAAUGGCGACCAAUAAAUUAAACAGCACGUAAUUGCCAAAAGUCAUUAGAGCCACAAAAUACAAUGCGGCCCAAUGACUUGUCUUUUCCAU ..........(((..((...............((((((.......................)))))).((((((((.(.(((.((........)).)))).)))))))).))..)))... ( -20.80) >DroAna_CAF1 9962 120 + 1 CUCUACCUCUGAACUGAAUCCCUCAACCAAAAUGGCCACCAAUAAAUUAAAUAGCACAUAAUUGCCGAACGUCAUUAGGGCCACAAAGUACAAUGCGGCCCAAUGACUUGUUUUUUCCAU ...............................((((.....(((((........(((......))).....((((((.(((((.((........)).))))))))))))))))....)))) ( -23.00) >DroPer_CAF1 25339 120 + 1 CUCUACCUCUGAACUGAAUCCCUCAACCAAAAUGGCCACCAAUAAAUUAAAUAGCACAUAAUUGCCGAAUGUCAUUAGGGCCACAAAGUACAAUGCGGCCCAAUGACUAGUCUUUUCCAU ...............................((((..................(((......))).((..((((((.(((((.((........)).)))))))))))...))....)))) ( -23.70) >consensus CUCUACCUCUGAACUGAAUCCCUCAACCAAAAUGGCCACCAAUAAAUUAAAUAGCACAUAAUUGCCGAACGUCAUUAGGGCCACAAAGUACAAUGCGGCCCAAUGACUUGUCUUUUCCAU ..........(((..((...............((((...........................))))...((((((.(((((.((........)).)))))))))))...))..)))... (-19.92 = -19.98 + 0.06)


| Location | 5,960,646 – 5,960,766 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -32.94 |
| Energy contribution | -32.97 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
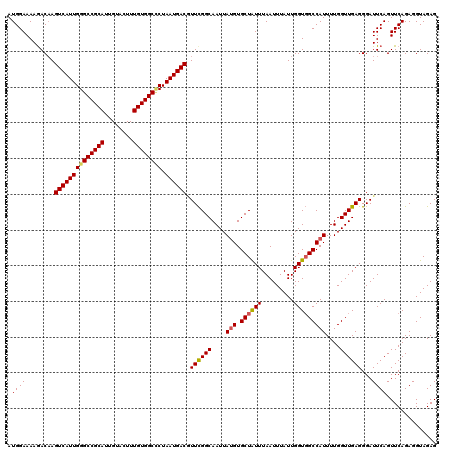
>X_DroMel_CAF1 5960646 120 - 22224390 AUGGAAAAGACAAGUCAUUGGGCCGCAUUGUACUUUGUGGCACUAAUGACGUUCGGCAAUUAUGUGCUAUUUAAUUUAUUGGUGGCCAUUUUGGUUGAGGGAUUCAGUUCAGAGGUAGAG ...(((..((...((((((((((((((........)))))).)))))))).((((((....(((.((((((.........)))))))))....))))))....))..))).......... ( -35.80) >DroGri_CAF1 19170 120 - 1 AUGGAAAAGACAAGUCAUUGGGCCGCACUGUACUUUGUGGCCCUAAUGACGUUCGGCAAUUAUGUGCUGUUUAAUUUAUUGGUGGCCAUUUUGGUUGAGGGAUUCAGUUCAGAGGUAGAG ...(((..((...((((((((((((((........)))))))).)))))).((((((....(((.((..((.........))..)))))....))))))....))..))).......... ( -35.80) >DroWil_CAF1 8749 120 - 1 AUGGAAAAGACAAGUCAUUGGGCCGCAUUGUAUUUUGUGGCCCUAAUGACAUUCGGCAAUUAUGUGCUAUUUAAUUUAUUGGUGGCCAUUUUGGUUGAGGGAUUCAGUUCAGAGGUAGAG ...(((..((...((((((((((((((........)))))))).)))))).((((((....(((.((((((.........)))))))))....))))))....))..))).......... ( -38.20) >DroMoj_CAF1 22381 120 - 1 AUGGAAAAGACAAGUCAUUGGGCCGCAUUGUAUUUUGUGGCUCUAAUGACUUUUGGCAAUUACGUGCUGUUUAAUUUAUUGGUCGCCAUUUUGGUUGAGGGAUUCAGUUCAGAGGUAGAG ((((....(((((((((((((((((((........)))))).))))))))))..((((......)))).............))).)))).....(((((........)))))........ ( -32.90) >DroAna_CAF1 9962 120 - 1 AUGGAAAAAACAAGUCAUUGGGCCGCAUUGUACUUUGUGGCCCUAAUGACGUUCGGCAAUUAUGUGCUAUUUAAUUUAUUGGUGGCCAUUUUGGUUGAGGGAUUCAGUUCAGAGGUAGAG .((((........((((((((((((((........)))))))).)))))).((((((....(((.((((((.........)))))))))....))))))...)))).............. ( -37.40) >DroPer_CAF1 25339 120 - 1 AUGGAAAAGACUAGUCAUUGGGCCGCAUUGUACUUUGUGGCCCUAAUGACAUUCGGCAAUUAUGUGCUAUUUAAUUUAUUGGUGGCCAUUUUGGUUGAGGGAUUCAGUUCAGAGGUAGAG ...(((..((...((((((((((((((........)))))))).)))))).((((((....(((.((((((.........)))))))))....))))))....))..))).......... ( -38.20) >consensus AUGGAAAAGACAAGUCAUUGGGCCGCAUUGUACUUUGUGGCCCUAAUGACGUUCGGCAAUUAUGUGCUAUUUAAUUUAUUGGUGGCCAUUUUGGUUGAGGGAUUCAGUUCAGAGGUAGAG .((((........((((((((((((((........)))))))).)))))).((((((....(((.((((((.........)))))))))....))))))...)))).............. (-32.94 = -32.97 + 0.03)


| Location | 5,960,686 – 5,960,806 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -30.79 |
| Energy contribution | -30.98 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5960686 120 - 22224390 ACUGACGCAAGAGGAUUGGAACGUCGUCCUGUUCAACGGAAUGGAAAAGACAAGUCAUUGGGCCGCAUUGUACUUUGUGGCACUAAUGACGUUCGGCAAUUAUGUGCUAUUUAAUUUAUU ......(((....((((((((((((.(((.((((....)))))))...)))..((((((((((((((........)))))).))))))))))))..)))))...)))............. ( -41.20) >DroVir_CAF1 20378 120 - 1 UCUGACGCAAGAGGAUUGGAACGUGGUUCUAUUCAAUGGCAUGGAAAAGACAAGUCAUUGGGCCGCAUUGUACUUUGUGGCUCUAAUGACGUUCGGCAAUUAUGUGCUAUUUAAUUUAUU .....(....)..((.((((((...)))))).))(((((((((.....((...((((((((((((((........)))))).))))))))..))........)))))))))......... ( -35.82) >DroGri_CAF1 19210 120 - 1 ACUGACGCAAGAGGAUUGGAACGUGGUUCUAUUCAAUGGCAUGGAAAAGACAAGUCAUUGGGCCGCACUGUACUUUGUGGCCCUAAUGACGUUCGGCAAUUAUGUGCUGUUUAAUUUAUU ...((((((....(((((((((((..((((((.(....).))))))...))..((((((((((((((........)))))))).))))))))))..)))))...))).)))......... ( -37.60) >DroWil_CAF1 8789 120 - 1 ACUCACACAAGAGGAUUGGAACGUCGUCUUGUUCAACGGCAUGGAAAAGACAAGUCAUUGGGCCGCAUUGUAUUUUGUGGCCCUAAUGACAUUCGGCAAUUAUGUGCUAUUUAAUUUAUU .(((......)))(((((((.....(((((.((((......)))).)))))..((((((((((((((........)))))))).))))))....((((......)))).))))))).... ( -38.50) >DroMoj_CAF1 22421 120 - 1 ACUAACGCAAGAGGAUUGGAACGUUGUCCUAUUCAAUGGCAUGGAAAAGACAAGUCAUUGGGCCGCAUUGUAUUUUGUGGCUCUAAUGACUUUUGGCAAUUACGUGCUGUUUAAUUUAUU .....(....).((((.(((......))).))))(((((((((.......(((((((((((((((((........)))))).)))))))))..)).......)))))))))......... ( -38.04) >DroAna_CAF1 10002 120 - 1 UCUAACACAAGAGGAUUGGAACGUCGUACUCUUCAAUGGAAUGGAAAAAACAAGUCAUUGGGCCGCAUUGUACUUUGUGGCCCUAAUGACGUUCGGCAAUUAUGUGCUAUUUAAUUUAUU .....((((....((((((((((((((.......(((((..((.......))..)))))((((((((........))))))))..)))))))))..))))).)))).............. ( -36.70) >consensus ACUGACGCAAGAGGAUUGGAACGUCGUCCUAUUCAAUGGCAUGGAAAAGACAAGUCAUUGGGCCGCAUUGUACUUUGUGGCCCUAAUGACGUUCGGCAAUUAUGUGCUAUUUAAUUUAUU ............((((((((((...(((...((((......))))...)))..((((((((((((((........)))))))).))))))))))((((......))))...))))))... (-30.79 = -30.98 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:45 2006