
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,940,487 – 5,940,628 |
| Length | 141 |
| Max. P | 0.995749 |

| Location | 5,940,487 – 5,940,588 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -20.56 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5940487 101 - 22224390 GCUCAACCAGUCUCUUCACAUGGCAACAGAUGGUAGAGAUUGCUUUUUUAUUUGGAAGAGAUAUUCCCCUAGAA---------GCACUGAUUUGGGGUCACCUUUUCGAA ..((...(((((((((((..((....))..))).))))))))............((((((......(((((((.---------.......)))))))....)))))))). ( -27.00) >DroSec_CAF1 27885 106 - 1 UUUCAUCCAGUGUCUUCACAUGGCAACAGAUGGAAAAGAUUGAUUUUUCAAUUGGUGAAGACAUCC----AUUUGGGGUAACCCCACUGAUUUGGGGUCACCUUUCGGUA ......((.((((((((((.((....))..(((((((......)))))))....))))))))))..----....(((((.((((((......)))))).)))))..)).. ( -37.30) >DroSim_CAF1 34163 110 - 1 GUUCAACCAGUGUCUUCACAUGGCAACGGAUGGAAAAGAUUGAUUUUUCAAUUGGUGAAGAUAUCCACCUAUUUGGGGUCACCCCACUGAUUUGGGGUCACCUUUCGGUA .....(((.((((((((((.((....))..(((((((......)))))))....))))))))))(((......)))(((.((((((......)))))).)))....))). ( -36.70) >consensus GUUCAACCAGUGUCUUCACAUGGCAACAGAUGGAAAAGAUUGAUUUUUCAAUUGGUGAAGAUAUCC_CCUAUUUGGGGU_ACCCCACUGAUUUGGGGUCACCUUUCGGUA .........((((((((((.((....))..(((((((......)))))))....)))))))))).........((((.....))))((((...(((....))).)))).. (-20.56 = -22.10 + 1.54)


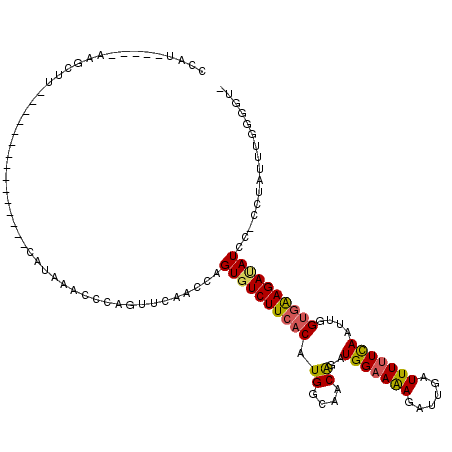
| Location | 5,940,514 – 5,940,628 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.65 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5940514 114 - 22224390 CAUUAUUUAAAGCUUUGGUUGGAUUGUAACCACAAGCCCAGCUCAACCAGUCUCUUCACAUGGCAACAGAUGGUAGAGAUUGCUUUUUUAUUUGGAAGAGAUAUUCCCCUAGAA------ ........(((((.((((((((.((((....))))..))))).)))..((((((((((..((....))..))).)))))))))))).......((((......)))).......------ ( -29.20) >DroSec_CAF1 27915 96 - 1 CCAU-----AAGCUU---------------CAUAAACAAAUUUCAUCCAGUGUCUUCACAUGGCAACAGAUGGAAAAGAUUGAUUUUUCAAUUGGUGAAGACAUCC----AUUUGGGGUA ....-----......---------------.........((..((....((((((((((.((....))..(((((((......)))))))....))))))))))..----...))..)). ( -24.70) >DroSim_CAF1 34193 100 - 1 CCAU-----AAGCUU---------------CAUAAACCCAGUUCAACCAGUGUCUUCACAUGGCAACGGAUGGAAAAGAUUGAUUUUUCAAUUGGUGAAGAUAUCCACCUAUUUGGGGUC ....-----......---------------......(((((........((((((((((.((....))..(((((((......)))))))....))))))))))........)))))... ( -25.59) >consensus CCAU_____AAGCUU_______________CAUAAACCCAGUUCAACCAGUGUCUUCACAUGGCAACAGAUGGAAAAGAUUGAUUUUUCAAUUGGUGAAGAUAUCC_CCUAUUUGGGGU_ .................................................((((((((((.((....))..(((((((......)))))))....))))))))))................ (-14.52 = -14.87 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:30 2006