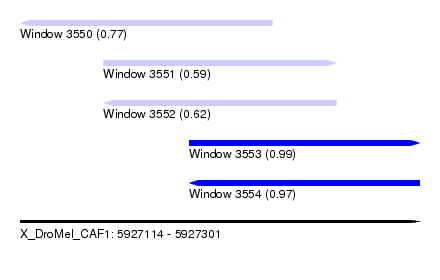
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,927,114 – 5,927,301 |
| Length | 187 |
| Max. P | 0.990806 |

| Location | 5,927,114 – 5,927,232 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5927114 118 - 22224390 -UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUUCAAGCAUUAAAUUUACUUAUAAUAUGCUGUGCUUAAUUGUGGAGAAGUAAUAAUUAACACCUAGUUGAAGGUGCAG-UCCCUCG -(((((.((.((((((((....))))))))....(((((((((...((......))...)))))(((.((((((((.........))))))))))).....)))))))))))-....... ( -25.00) >DroSec_CAF1 14812 118 - 1 -UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAGCGUUGAAUUUACUUAUAAUAUUCUGUGCUUAAUUAUGGAGAAGUAAUAAUUAACACCUAGUUGUAGGUGCAG-UCCCUCC -(((((....((((((((....))))))))....)))))(((((((..((((((......(((((((......)))))))))))))...)))))(((((....)))))))..-....... ( -26.90) >DroSim_CAF1 18948 118 - 1 -UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAGCGUUGAAUUUACUUAUAAUAUUCUGUGCUUGAUUAUGGAGAAGUAAUAAUUAACACCUAGUUGUAGGUGCAG-UCCCUCU -(((((....((((((((....))))))))....)))))(((((((..((((((...(((.((.......)).)))....))))))...)))))(((((....)))))))..-....... ( -28.10) >DroEre_CAF1 14318 119 - 1 -UUGCAGCCACCUAAUUAUCGUUAAUUGGGCAUAUGCAAGCAUUGAAUUUUCUUAUAAUAUUCUGUGCUUAAUUCUGGCGAAGUAAUAAUUAGCAUCUAAUUCUAGGUGCAGCUACUUCG -.....((((((((((((....)))))))).......((((((.((((...........)))).)))))).....))))((((((.......(((((((....)))))))...)))))). ( -33.50) >DroYak_CAF1 16907 120 - 1 AUUACAGCUACCUAAUUAUAGUUAAUUGGGAAUAUGCAAGCAUUGAAUUCUCCUAUAAUAUUCUGUGCUUAAUUAUGGAGUAUUAACAAUUAACACCUAAUUCUAGGUGCAGGUUCCUCG .....((..((((.......((((((((((((((((....)))...)))))....((((((((((((......)))))))))))).))))))))(((((....)))))..))))..)).. ( -28.20) >consensus _UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAGCAUUGAAUUUACUUAUAAUAUUCUGUGCUUAAUUAUGGAGAAGUAAUAAUUAACACCUAGUUGUAGGUGCAG_UCCCUCG .((((..(((((((((((....)))))))).......((((((.((((...........)))).)))))).....)))....)))).......((((((....))))))........... (-20.82 = -20.90 + 0.08)



| Location | 5,927,153 – 5,927,262 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -6.99 |
| Energy contribution | -7.71 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5927153 109 + 22224390 CUCCACAAUUAAGCACAGCAUAUUAUAAGUAAAUUUAAUGCUUGAAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA-UUGC----------AUAUACCUAUUAUACACAUUUGCAAC ............(((....(((((.((((((.......)))))))))))...........((((..((((((..(((..-..))----------)..))))))...))))....)))... ( -18.80) >DroSec_CAF1 14851 109 + 1 CUCCAUAAUUAAGCACAGAAUAUUAUAAGUAAAUUCAACGCUUGCAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA-UUGC----------AUUUACCUAAUAUACAAACUUGCAAC ............(((..((((((..(((((.........))))).)))))).........(((((((((((((.(((..-..))----------).))))))))).))))....)))... ( -22.70) >DroSim_CAF1 18987 109 + 1 CUCCAUAAUCAAGCACAGAAUAUUAUAAGUAAAUUCAACGCUUGCAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA-UUGC----------AUUUACCUAAUAUACAAACUUGCAAC ............(((..((((((..(((((.........))))).)))))).........(((((((((((((.(((..-..))----------).))))))))).))))....)))... ( -22.70) >DroEre_CAF1 14358 103 + 1 CGCCAGAAUUAAGCACAGAAUAUUAUAAGAAAAUUCAAUGCUUGCAUAUGCCCAAUUAACGAUAAUUAGGUGGCUGCAA-GUAU------ACAUGUAUAACUAAUAA----------AUG ........(((.(((..((((.((.....)).)))).(((((((((..((((.(((((....))))).))))..)))))-))))------...))).))).......----------... ( -18.40) >DroYak_CAF1 16947 120 + 1 CUCCAUAAUUAAGCACAGAAUAUUAUAGGAGAAUUCAAUGCUUGCAUAUUCCCAAUUAACUAUAAUUAGGUAGCUGUAAUGUAUGUACGCACAUAUGUACCUAAAAUACUAAUUUCCAAG (((((((((............))))).)))).......(((.(((((((...((.(((.(((....))).))).))....))))))).)))............................. ( -17.20) >consensus CUCCAUAAUUAAGCACAGAAUAUUAUAAGUAAAUUCAAUGCUUGCAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA_UUGC__________AUAUACCUAAUAUACAAACUUGCAAC .........((((((..((((...........))))..))))))....................((((((((.........................))))))))............... ( -6.99 = -7.71 + 0.72)

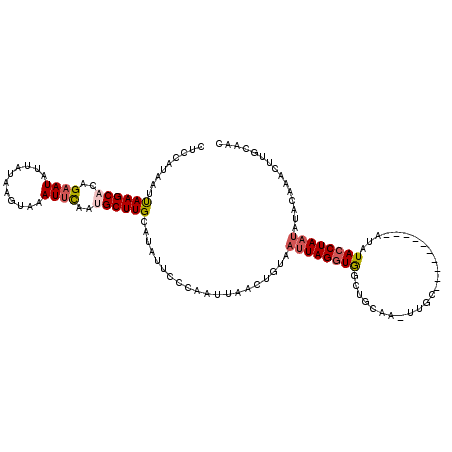

| Location | 5,927,153 – 5,927,262 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5927153 109 - 22224390 GUUGCAAAUGUGUAUAAUAGGUAUAU----------GCAA-UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUUCAAGCAUUAAAUUUACUUAUAAUAUGCUGUGCUUAAUUGUGGAG (((((((.((((((((.....)))))----------))).-)))))))..((((((((....)))))))).......((((((...((...........))...)))))).......... ( -25.70) >DroSec_CAF1 14851 109 - 1 GUUGCAAGUUUGUAUAUUAGGUAAAU----------GCAA-UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAGCGUUGAAUUUACUUAUAAUAUUCUGUGCUUAAUUAUGGAG ....((((((((((((((.(((...(----------((..-..)))))).((((((((....))))))))))))))))))).)))................((((((......)))))). ( -26.50) >DroSim_CAF1 18987 109 - 1 GUUGCAAGUUUGUAUAUUAGGUAAAU----------GCAA-UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAGCGUUGAAUUUACUUAUAAUAUUCUGUGCUUGAUUAUGGAG ....(((((..((((..(((((((((----------.(((-(((((....((((((((....))))))))....))))...)))).)))))))))..)))).....)))))......... ( -26.70) >DroEre_CAF1 14358 103 - 1 CAU----------UUAUUAGUUAUACAUGU------AUAC-UUGCAGCCACCUAAUUAUCGUUAAUUGGGCAUAUGCAAGCAUUGAAUUUUCUUAUAAUAUUCUGUGCUUAAUUCUGGCG ...----------..............(((------(...-.))))((((((((((((....)))))))).......((((((.((((...........)))).)))))).....)))). ( -22.80) >DroYak_CAF1 16947 120 - 1 CUUGGAAAUUAGUAUUUUAGGUACAUAUGUGCGUACAUACAUUACAGCUACCUAAUUAUAGUUAAUUGGGAAUAUGCAAGCAUUGAAUUCUCCUAUAAUAUUCUGUGCUUAAUUAUGGAG (((.(.((((.((((..((.((((....)))).)).))))......((..((((((((....)))))))).....))((((((.((((...........)))).)))))))))).).))) ( -23.70) >consensus GUUGCAAAUUUGUAUAUUAGGUAAAU__________GCAA_UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAGCAUUGAAUUUACUUAUAAUAUUCUGUGCUUAAUUAUGGAG ...............................................(((((((((((....)))))))).......((((((.((((...........)))).)))))).....))).. (-14.52 = -14.52 + -0.00)

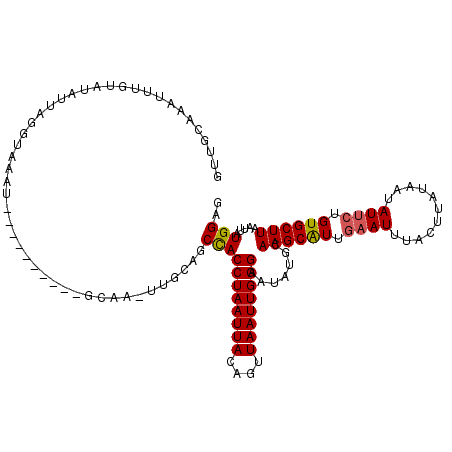

| Location | 5,927,193 – 5,927,301 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.18 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -9.35 |
| Energy contribution | -9.95 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5927193 108 + 22224390 CUUGAAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA-UUGC----------AUAUACCUAUUAUACACAUUUGCAACAAAAAAUCCGGU-UUCCACUUUGUGCAAAAAUAAAUAUUU ...(((((((..........((((..((((((..(((..-..))----------)..))))))...))))..((((((.((((.........-......))))))))))....))))))) ( -19.56) >DroSec_CAF1 14891 107 + 1 CUUGCAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA-UUGC----------AUUUACCUAAUAUACAAACUUGCAACAAGAAAUCCGGU-UUCCACUUUGUGCAAAACUAUAUAUA- .(((((((.........(((((..(((((((((.(((..-..))----------).))))))))).......((((...)))).....))))-).......)))))))...........- ( -24.09) >DroSim_CAF1 19027 101 + 1 CUUGCAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA-UUGC----------AUUUACCUAAUAUACAAACUUGCAACAAAAAAUCCGGU-UCCCACUUUGUGCAAAA------AUA- .(((((((............(((((((((((((.(((..-..))----------).))))))))).))))...................((.-..))....)))))))..------...- ( -22.10) >DroEre_CAF1 14398 98 + 1 CUUGCAUAUGCCCAAUUAACGAUAAUUAGGUGGCUGCAA-GUAU------ACAUGUAUAACUAAUAA----------AUGGGAAAAUCCGGUUUUCCG--CUAUGCAAAAAUAU--AUA- ((((((..((((.(((((....))))).))))..)))))-)...------...((((((.(....((----------((.((.....)).))))...)--.)))))).......--...- ( -21.10) >DroYak_CAF1 16987 117 + 1 CUUGCAUAUUCCCAAUUAACUAUAAUUAGGUAGCUGUAAUGUAUGUACGCACAUAUGUACCUAAAAUACUAAUUUCCAAGAAAAAAUCCGGAUUUCCCCCCAAUGCAAAAAUUU--AUA- .((((((..................((((((.......((((((((....)))))))))))))).........................((........)).))))))......--...- ( -15.81) >consensus CUUGCAUAUUCCCAAUUAACUGUAAUUAGGUGGCUGCAA_UUGC__________AUAUACCUAAUAUACAAACUUGCAACAAAAAAUCCGGU_UUCCACUUUGUGCAAAAAUAU__AUA_ .(((((((................((((((((.........................))))))))........................((....))....)))))))............ ( -9.35 = -9.95 + 0.60)

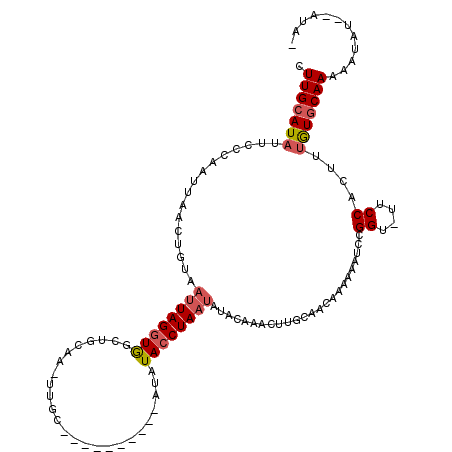
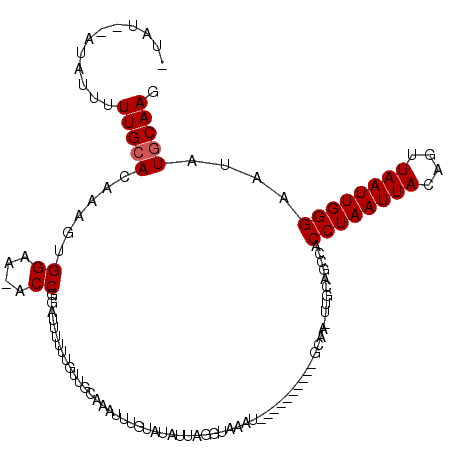
| Location | 5,927,193 – 5,927,301 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.18 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -11.48 |
| Energy contribution | -11.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5927193 108 - 22224390 AAAUAUUUAUUUUUGCACAAAGUGGAA-ACCGGAUUUUUUGUUGCAAAUGUGUAUAAUAGGUAUAU----------GCAA-UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUUCAAG .........((((..(.....)..)))-)..((((.....(((((((.((((((((.....)))))----------))).-)))))))..((((((((....))))))))...))))... ( -25.90) >DroSec_CAF1 14891 107 - 1 -UAUAUAUAGUUUUGCACAAAGUGGAA-ACCGGAUUUCUUGUUGCAAGUUUGUAUAUUAGGUAAAU----------GCAA-UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAG -..........((((((((....((((-(.....))))))).)))))).(((((((((.(((...(----------((..-..)))))).((((((((....))))))))))))))))). ( -25.50) >DroSim_CAF1 19027 101 - 1 -UAU------UUUUGCACAAAGUGGGA-ACCGGAUUUUUUGUUGCAAGUUUGUAUAUUAGGUAAAU----------GCAA-UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAG -...------.((((((((((((((..-.)))....))))).)))))).(((((((((.(((...(----------((..-..)))))).((((((((....))))))))))))))))). ( -26.10) >DroEre_CAF1 14398 98 - 1 -UAU--AUAUUUUUGCAUAG--CGGAAAACCGGAUUUUCCCAU----------UUAUUAGUUAUACAUGU------AUAC-UUGCAGCCACCUAAUUAUCGUUAAUUGGGCAUAUGCAAG -..(--((((...((.((((--(((....))((......))..----------......))))).)).))------)))(-(((((....((((((((....))))))))....)))))) ( -24.00) >DroYak_CAF1 16987 117 - 1 -UAU--AAAUUUUUGCAUUGGGGGGAAAUCCGGAUUUUUUCUUGGAAAUUAGUAUUUUAGGUACAUAUGUGCGUACAUACAUUACAGCUACCUAAUUAUAGUUAAUUGGGAAUAUGCAAG -...--......((((((..((..(((((....)))))..))..)......((((..((.((((....)))).)).))))..........((((((((....))))))))....))))). ( -28.00) >consensus _UAU__AUAUUUUUGCACAAAGUGGAA_ACCGGAUUUUUUGUUGCAAAUUUGUAUAUUAGGUAAAU__________GCAA_UUGCAGCCACCUAAUUACAGUUAAUUGGGAAUAUGCAAG ............(((((......((....))...........................................................((((((((....))))))))....))))). (-11.48 = -11.68 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:21 2006