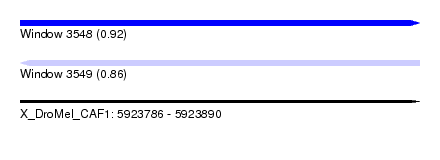
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,923,786 – 5,923,890 |
| Length | 104 |
| Max. P | 0.923410 |

| Location | 5,923,786 – 5,923,890 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
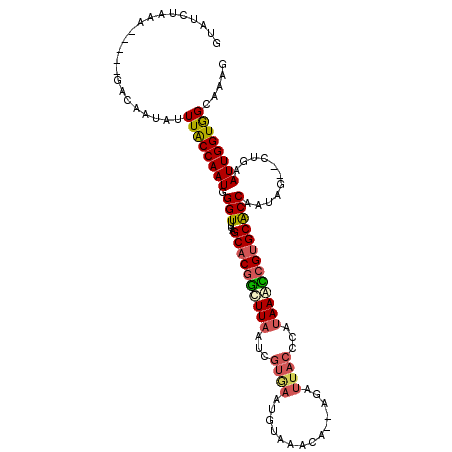
>X_DroMel_CAF1 5923786 104 + 22224390 GUAUCUAAA-----CACAAUAUUUACCAAUGGGUUUUAGCACGGCUUAAUCGUGAAUGUAAACA--AGAUUACCCAUAAGCCGUGCACCAAUUG--CUUAAUUGGUAGCAAAG .........-----........((((((((((((....((((((((((...((((.((....))--...))))...)))))))))).......)--))).))))))))..... ( -27.10) >DroSec_CAF1 10399 104 + 1 GUAUUUAAA-----GACAAUAUUUACCAAUGGGUUUUAGCACGGUUUUAUCGUGAAUGUAAACA--AGAUUACCCAUAAACCGUGCACCUAUAG--CUGAAUUGGUAGCAAAG .........-----........((((((((.((((.(((((((((((....((((.((....))--...))))....)))))))))...)).))--))..))))))))..... ( -24.60) >DroSim_CAF1 11449 104 + 1 GUAUUUAAA-----GACAAUAUUUACCAAUGGGUUUUAGCACGGUUUAAUCGUGAAUGUAAACA--AGAUUACCCAUAAACCGUGCACCAAUAG--CUGAAUUGGUGGCAAAG .........-----........((((((((.((((...((((((((((...((((.((....))--...))))...))))))))))......))--))..))))))))..... ( -25.10) >DroEre_CAF1 10996 94 + 1 GAAUCGAAA-----GACAAUAUUUGCCAAUGGGUUUAGGCUCAGCUUAA------------GCAACAGAUUAACCAUAAAUCGUGCGCCACCAG--CUAGAUUGGUGGCAAAG ...((....-----))........(((.....((((((((...))))))------------))....((((.......))))).))((((((((--.....)))))))).... ( -26.10) >DroYak_CAF1 13733 111 + 1 GAAUCGAAAGAAAAGACAAUAUUUACCAAUGGGUUUUAGCACGACUUAAUCGUAAGUGUAAGCA--AAAUUACCCAUAACUCGUGCACCAAUAGCACUGAAUUGGUGGCAAAG ...((....))...........(((((((((((((((.((((.(((((....)))))))..)).--)))..)))).......((((.......))))...))))))))..... ( -23.80) >consensus GUAUCUAAA_____GACAAUAUUUACCAAUGGGUUUUAGCACGGCUUAAUCGUGAAUGUAAACA__AGAUUACCCAUAAACCGUGCACCAAUAG__CUGAAUUGGUGGCAAAG ......................((((((((.(((....((((((((((...((((..............))))...)))))))))))))...........))))))))..... (-17.22 = -17.78 + 0.56)

| Location | 5,923,786 – 5,923,890 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.01 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5923786 104 - 22224390 CUUUGCUACCAAUUAAG--CAAUUGGUGCACGGCUUAUGGGUAAUCU--UGUUUACAUUCACGAUUAAGCCGUGCUAAAACCCAUUGGUAAAUAUUGUG-----UUUAGAUAC .(((((((((((((...--.)))))))((((((((((((.((((...--...))))...))....))))))))))...........)))))).......-----......... ( -27.30) >DroSec_CAF1 10399 104 - 1 CUUUGCUACCAAUUCAG--CUAUAGGUGCACGGUUUAUGGGUAAUCU--UGUUUACAUUCACGAUAAAACCGUGCUAAAACCCAUUGGUAAAUAUUGUC-----UUUAAAUAC ......(((((((...(--(.....))(((((((((((.(((((...--...)))).....).)).)))))))))........))))))).........-----......... ( -20.10) >DroSim_CAF1 11449 104 - 1 CUUUGCCACCAAUUCAG--CUAUUGGUGCACGGUUUAUGGGUAAUCU--UGUUUACAUUCACGAUUAAACCGUGCUAAAACCCAUUGGUAAAUAUUGUC-----UUUAAAUAC .((((((((((((....--..))))))((((((((((((.((((...--...))))...))....))))))))))...........)))))).......-----......... ( -27.30) >DroEre_CAF1 10996 94 - 1 CUUUGCCACCAAUCUAG--CUGGUGGCGCACGAUUUAUGGUUAAUCUGUUGC------------UUAAGCUGAGCCUAAACCCAUUGGCAAAUAUUGUC-----UUUCGAUUC ....(((((((......--.)))))))(.(((((.....((((((..(((((------------((.....))))...)))..))))))....))))))-----......... ( -22.00) >DroYak_CAF1 13733 111 - 1 CUUUGCCACCAAUUCAGUGCUAUUGGUGCACGAGUUAUGGGUAAUUU--UGCUUACACUUACGAUUAAGUCGUGCUAAAACCCAUUGGUAAAUAUUGUCUUUUCUUUCGAUUC .(((((((..(((((.((((.......)))))))))((((((..(((--.((....(((((....)))))...)).))))))))))))))))..................... ( -25.20) >consensus CUUUGCCACCAAUUCAG__CUAUUGGUGCACGGUUUAUGGGUAAUCU__UGUUUACAUUCACGAUUAAGCCGUGCUAAAACCCAUUGGUAAAUAUUGUC_____UUUAGAUAC .((((((((((((........))))))((((((((((............................))))))))))...........))))))..................... (-18.85 = -19.01 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:16 2006