
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,902,828 – 5,903,040 |
| Length | 212 |
| Max. P | 0.968170 |

| Location | 5,902,828 – 5,902,934 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -28.06 |
| Energy contribution | -27.75 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5902828 106 - 22224390 CAAACAACUGCAGCGGCGGCAAGAAAGCGGCAGCCAAAACAAAAGCAAAAAGGCCAACAUCUCUGACGGCCUUUCACCUGUUGGAUGUCGAGAAAAGCAACUGCCU .........((((..((.((......))((((.((((......((...(((((((..((....))..)))))))...)).)))).)))).......))..)))).. ( -29.90) >DroSec_CAF1 19480 106 - 1 CAAACAACUGCAGCGGCGGCAAGAAAGCGGCAGCCAAAACAAAAGCAAAAAGGCCAACAUCUCUGACGGCCUUUCACCUGUUGGAUGUCGGGAAAAGCAACUGCCU .........((((..((.((......))((((.((((......((...(((((((..((....))..)))))))...)).)))).)))).......))..)))).. ( -29.90) >DroSim_CAF1 16848 106 - 1 CAAACAACUGCAGCGGCGGCAAGAAAGCGGCAGCCAAAACAAAAGCAAAAAGGCCAACAUCUCUGACGGCCUUUCACCUGUUGGAUGUCGAGAAAAGCAACUGCCU .........((((..((.((......))((((.((((......((...(((((((..((....))..)))))))...)).)))).)))).......))..)))).. ( -29.90) >DroEre_CAF1 19498 105 - 1 CAAACAACUGCAGCAGCGGCAAGAAAGUGGCAGCCAAAACAAAAGCGAAGAGGCCAACGUCUCUGACGGCCUUUCACCUGUUGAAUGUCGAAA-AAGCAACUGCCU ............((((..((.......(((((.....((((...(.((.((((((...(((...))))))))))).).))))...)))))...-..))..)))).. ( -24.90) >consensus CAAACAACUGCAGCGGCGGCAAGAAAGCGGCAGCCAAAACAAAAGCAAAAAGGCCAACAUCUCUGACGGCCUUUCACCUGUUGGAUGUCGAGAAAAGCAACUGCCU .........((((..((.((......))((((.((((......((...(((((((..((....))..)))))))...)).)))).)))).......))..)))).. (-28.06 = -27.75 + -0.31)

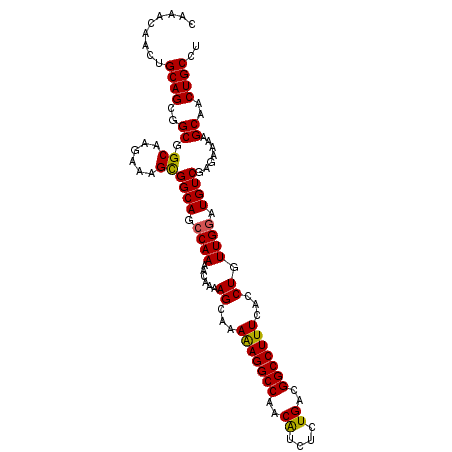
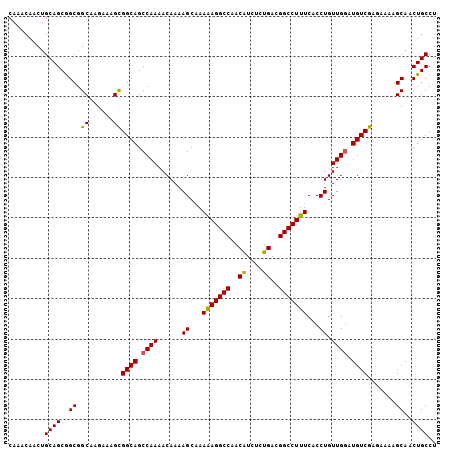
| Location | 5,902,903 – 5,903,005 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 96.59 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -27.09 |
| Energy contribution | -27.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5902903 102 + 22224390 GCCGCUUUCUUGCCGCCGCUGCAGUUGUUUGUCGAAACUUAAUUGUAACUGCCAAAUAAUUCGUGGCGGCCACAGAAAGAGAUUAUCAUUAUUAUUACUAUU- ..(.((((((.((((((((.(.(((((((((.((..((......))...)).))))))))))))))))))...)))))).).....................- ( -28.70) >DroSec_CAF1 19555 102 + 1 GCCGCUUUCUUGCCGCCGCUGCAGUUGUUUGUCGAAACUUAAUUGUAACUGCCAAAUAAUUCGUGGCGGCCACAGAAAGAGAUUAUCAUUAUUAUUACUAUU- ..(.((((((.((((((((.(.(((((((((.((..((......))...)).))))))))))))))))))...)))))).).....................- ( -28.70) >DroSim_CAF1 16923 102 + 1 GCCGCUUUCUUGCCGCCGCUGCAGUUGUUUGUCGAAACUUAAUUGUAACUGCCAAAUAAUUCGUGGCGGCCACAGAAAGAGAUUAUCAUUAUUAUUACUAUU- ..(.((((((.((((((((.(.(((((((((.((..((......))...)).))))))))))))))))))...)))))).).....................- ( -28.70) >DroEre_CAF1 19572 103 + 1 GCCACUUUCUUGCCGCUGCUGCAGUUGUUUGUCGAAACUUAAUUGUAACUGCCAAAUAAUUCGUGGCGGCCACAGAAACAGAUUAUCAUUAUCAUUGUUAUUA .....(((((.(((((..(.(.(((((((((.((..((......))...)).)))))))))))..)))))...)))))..(((.......))).......... ( -22.70) >consensus GCCGCUUUCUUGCCGCCGCUGCAGUUGUUUGUCGAAACUUAAUUGUAACUGCCAAAUAAUUCGUGGCGGCCACAGAAAGAGAUUAUCAUUAUUAUUACUAUU_ ..(.((((((.((((((((.(.(((((((((.((..((......))...)).))))))))))))))))))...)))))).)...................... (-27.09 = -27.15 + 0.06)

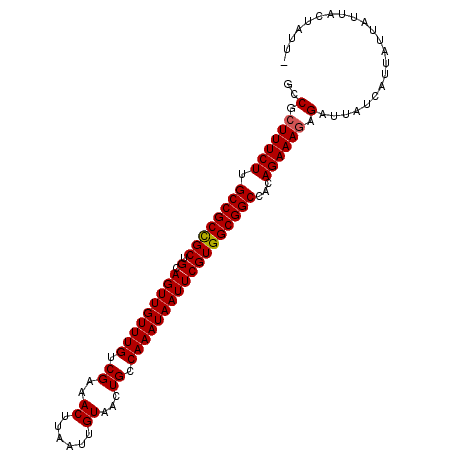
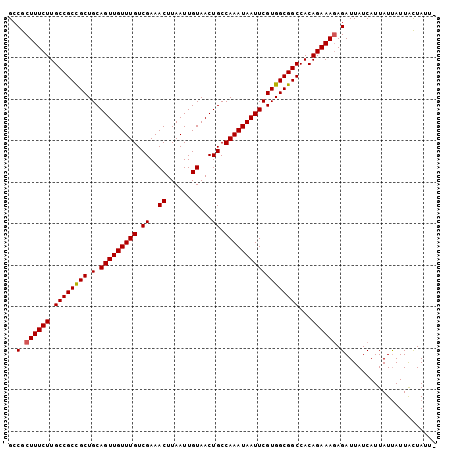
| Location | 5,902,903 – 5,903,005 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 96.59 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.85 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5902903 102 - 22224390 -AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGACAAACAACUGCAGCGGCGGCAAGAAAGCGGC -....................((.((((((...((((((.(((.....)))((((((........((((....))))))))))...)))))).)))))).)). ( -27.10) >DroSec_CAF1 19555 102 - 1 -AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGACAAACAACUGCAGCGGCGGCAAGAAAGCGGC -....................((.((((((...((((((.(((.....)))((((((........((((....))))))))))...)))))).)))))).)). ( -27.10) >DroSim_CAF1 16923 102 - 1 -AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGACAAACAACUGCAGCGGCGGCAAGAAAGCGGC -....................((.((((((...((((((.(((.....)))((((((........((((....))))))))))...)))))).)))))).)). ( -27.10) >DroEre_CAF1 19572 103 - 1 UAAUAACAAUGAUAAUGAUAAUCUGUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGACAAACAACUGCAGCAGCGGCAAGAAAGUGGC .........................(((((...(((((..(((.....)))((((((........((((....))))))))))....))))).)))))..... ( -19.40) >consensus _AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGACAAACAACUGCAGCGGCGGCAAGAAAGCGGC .....................((.((((((...((((((.(((.....)))((((((........((((....))))))))))...)))))).)))))).)). (-24.35 = -24.85 + 0.50)

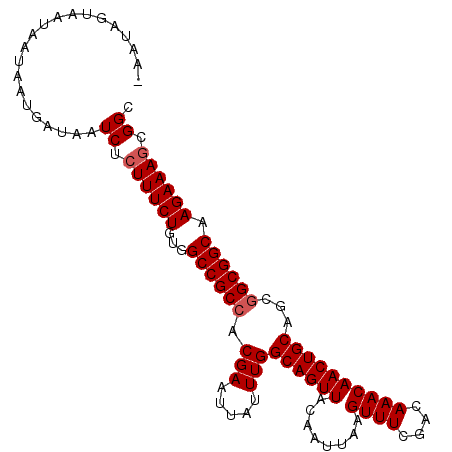

| Location | 5,902,934 – 5,903,040 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.04 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -23.51 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5902934 106 - 22224390 UUCCUUUCGACAGUUAAUAACAAUAGAAAGAGAGU------AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGA ......((((.(.(((((......(((((((((..------.....((....)).......)))))))))(((((.((((.........)))).))))).))))).).)))) ( -25.44) >DroSec_CAF1 19586 106 - 1 UUCCUUUCGACAGUUAAUAACAAUAGAAAGAGAGU------AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGA ......((((.(.(((((......(((((((((..------.....((....)).......)))))))))(((((.((((.........)))).))))).))))).).)))) ( -25.44) >DroSim_CAF1 16954 106 - 1 UUCCUUUCGACAGUUAAUAACAAUAGAAAGAGAGU------AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGA ......((((.(.(((((......(((((((((..------.....((....)).......)))))))))(((((.((((.........)))).))))).))))).).)))) ( -25.44) >DroEre_CAF1 19603 112 - 1 UUCCUUUCGACAGUUAAUGACAAUAGAAAGAGCGACAUAAUAAUAACAAUGAUAAUGAUAAUCUGUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGA ......((((.(.(((((......(((((.((...(((.((.((....)).)).))).....)).)))))(((((.((((.........)))).))))).))))).).)))) ( -18.60) >consensus UUCCUUUCGACAGUUAAUAACAAUAGAAAGAGAGU______AAUAGUAAUAAUAAUGAUAAUCUCUUUCUGUGGCCGCCACGAAUUAUUUGGCAGUUACAAUUAAGUUUCGA ......((((.(.(((((......(((((((((..........((....))..........)))))))))(((((.((((.........)))).))))).))))).).)))) (-23.51 = -23.82 + 0.31)

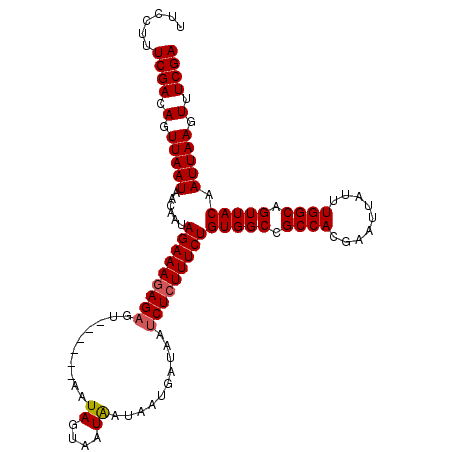
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:08 2006