
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,902,277 – 5,902,417 |
| Length | 140 |
| Max. P | 0.957408 |

| Location | 5,902,277 – 5,902,385 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.66 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -15.16 |
| Energy contribution | -16.72 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5902277 108 + 22224390 GGAAAAUGCCCAACUGGAUGCUGGGCAACUGAAAAGUAUACAGAUAUAUAUAUAUAUCUGCAUAUGUGCACAUGUGCAGAGAAAGUUAUGUCCACAAAUAAUUACUAC ......((((((.(.....).))))))(((....))).....((((((........((((((((((....))))))))))......))))))................ ( -30.94) >DroSec_CAF1 18944 99 + 1 GGAAAAUGCCCAACUGGAUGCUGGGCAACUGAAAAGUAUGCAUAUA-GUAUAUA--------UAUGUGCAUAUGUGCAGAGAAAGUCAUGUCCACAAAUAAUUACUAC (((...((((((.(.....).)))))).(((....(((((((((((-.......--------)))))))))))...)))...........)))............... ( -29.40) >DroSim_CAF1 16311 99 + 1 GGAAAAUGCCCAACUGGAUGCUGGGCAACUGAAAAGUAUGCAUAUA-GUAUAUA--------UAUGUGCAUAUGUGCAGAGAAAGUCAUGUCCACAAAUAAUUACUAC (((...((((((.(.....).)))))).(((....(((((((((((-.......--------)))))))))))...)))...........)))............... ( -29.40) >DroEre_CAF1 18955 92 + 1 GGAAAAUGCCCAACGGGAUGCUGGACAACUGAAAC-------UAUA-CCAUGU--------GCGCAUGCAUAUGUGCAGAGAAAGUCAUGUCCACAAAUAAUUACUAC ........(((...))).((.(((((((((....(-------(.((-(.((((--------((....))))))))).))....)))..))))))))............ ( -20.40) >DroYak_CAF1 19258 95 + 1 GGAAAAUGCCCAACUGGAAGCUGGACAACUGACUA-------UAUG-GCAUAC-----UACUUAUAUGCAUAUGUGCAGAGAAAGUCAUGUCCACAAAUAAUUACUAC .......((((....))..))((((((..((((((-------((((-(((((.-----......))))).)))))........))))))))))).............. ( -17.70) >consensus GGAAAAUGCCCAACUGGAUGCUGGGCAACUGAAAAGUAU_CAUAUA_GUAUAUA________UAUGUGCAUAUGUGCAGAGAAAGUCAUGUCCACAAAUAAUUACUAC (((...((((((.(.....).)))))).(((.........((((((.(((((((........))))))).)))))))))...........)))............... (-15.16 = -16.72 + 1.56)

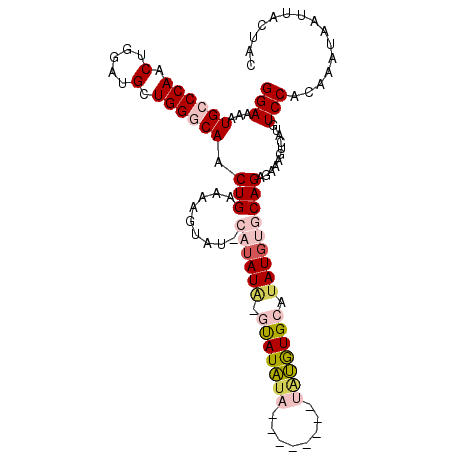
| Location | 5,902,277 – 5,902,385 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.66 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -18.00 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5902277 108 - 22224390 GUAGUAAUUAUUUGUGGACAUAACUUUCUCUGCACAUGUGCACAUAUGCAGAUAUAUAUAUAUAUCUGUAUACUUUUCAGUUGCCCAGCAUCCAGUUGGGCAUUUUCC ............((((.((((..............)))).))))((((((((((((....)))))))))))).........((((((((.....))))))))...... ( -32.34) >DroSec_CAF1 18944 99 - 1 GUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCACAUA--------UAUAUAC-UAUAUGCAUACUUUUCAGUUGCCCAGCAUCCAGUUGGGCAUUUUCC ((((((..(((.((((.(.(((............))).).)))).)--------))..)))-)))................((((((((.....))))))))...... ( -23.50) >DroSim_CAF1 16311 99 - 1 GUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCACAUA--------UAUAUAC-UAUAUGCAUACUUUUCAGUUGCCCAGCAUCCAGUUGGGCAUUUUCC ((((((..(((.((((.(.(((............))).).)))).)--------))..)))-)))................((((((((.....))))))))...... ( -23.50) >DroEre_CAF1 18955 92 - 1 GUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCAUGCGC--------ACAUGG-UAUA-------GUUUCAGUUGUCCAGCAUCCCGUUGGGCAUUUUCC ...............(((...((((..((.(((.(((.(((....))--------).))))-)).)-------)....))))(((((((.....)))))))....))) ( -20.70) >DroYak_CAF1 19258 95 - 1 GUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCAUAUAAGUA-----GUAUGC-CAUA-------UAGUCAGUUGUCCAGCUUCCAGUUGGGCAUUUUCC ............(((((.((((((((....((((....))))...)))).-----.)))))-))))-------........((((((((.....))))))))...... ( -23.00) >consensus GUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCACAUA________UAUAUAC_UAUAUG_AUACUUUUCAGUUGCCCAGCAUCCAGUUGGGCAUUUUCC ...(((..(((.((..((.(......).))..)).))))))........................................((((((((.....))))))))...... (-18.00 = -17.60 + -0.40)



| Location | 5,902,305 – 5,902,417 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.76 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -18.12 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5902305 112 - 22224390 UUU--------UCGGUUGCGUCUUCUUAGGCAGCCGAAAUGUAGUAAUUAUUUGUGGACAUAACUUUCUCUGCACAUGUGCACAUAUGCAGAUAUAUAUAUAUAUCUGUAUACUUUUCAG .((--------((((((((.(......).)))))))))).............((((.((((..............)))).))))((((((((((((....))))))))))))........ ( -33.24) >DroSec_CAF1 18972 103 - 1 UUU--------UCGGUUGCGUCUUCUUAGGCAGCCGAAAUGUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCACAUA--------UAUAUAC-UAUAUGCAUACUUUUCAG ..(--------((((((((.(......).)))))))))((((((((..(((.((((.(.(((............))).).)))).)--------))..)))-)))))............. ( -26.10) >DroSim_CAF1 16339 103 - 1 UUU--------UCGGUUGCGUCUUCUUAGGCAGCCGAAAUGUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCACAUA--------UAUAUAC-UAUAUGCAUACUUUUCAG ..(--------((((((((.(......).)))))))))((((((((..(((.((((.(.(((............))).).)))).)--------))..)))-)))))............. ( -26.10) >DroEre_CAF1 18983 97 - 1 UUUU-------UUGGUUGCGUCUUCUUGGGCAGCUGAAAUGUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCAUGCGC--------ACAUGG-UAUA-------GUUUCAG ..((-------(..(((((.((.....)))))))..)))....(.((((((.((..((.(......).))..))(((.(((....))--------).))).-.)))-------))).).. ( -19.50) >DroYak_CAF1 19286 107 - 1 UUUUCUGUGUUUCGGUUGCGUCUUCUUAGGCAGCUGAAAUGUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCAUAUAAGUA-----GUAUGC-CAUA-------UAGUCAG ....(((..((((((((((.(......).))))))))))..)))........(((((.((((((((....((((....))))...)))).-----.)))))-))))-------....... ( -29.80) >consensus UUU________UCGGUUGCGUCUUCUUAGGCAGCCGAAAUGUAGUAAUUAUUUGUGGACAUGACUUUCUCUGCACAUAUGCACAUA________UAUAUAC_UAUAUG_AUACUUUUCAG ...........((((((((.(......).)))))))).((((.(((..(((.((..((.(......).))..)).))))))))))................................... (-18.12 = -17.20 + -0.92)

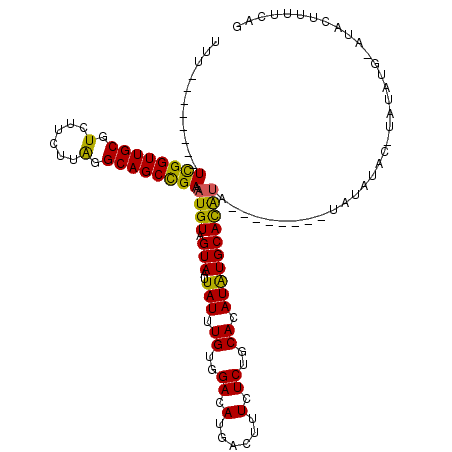

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:05 2006