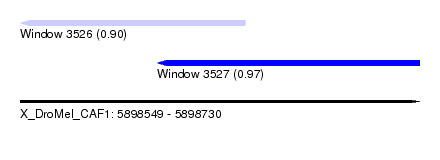
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,898,549 – 5,898,730 |
| Length | 181 |
| Max. P | 0.973881 |

| Location | 5,898,549 – 5,898,651 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.10 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5898549 102 - 22224390 UGCUAGUGGGUUAACUUGGCUAAAAGGCUUAUCCUAUGCCACAAAUGCCAGUCUUGGUA------------------GACUCGGUAGACCUGGUGGAAAACCCAACCAGUCCAAGAGUCG ((((((..(((.....((((....(((.....)))..)))).....)))....))))))------------------(((((....(((.((((((.....)).)))))))...))))). ( -31.00) >DroSec_CAF1 15281 102 - 1 UGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCCACAAAUGCCAGUCUUGGUA------------------GACUCGGUAGACCUGGUGGAAAACCCAACCAGUCCAAGAGUCG .((..(((((.((((((........)))))).)))))))......((((......))))------------------(((((....(((.((((((.....)).)))))))...))))). ( -31.40) >DroSim_CAF1 12588 102 - 1 UGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCCACAAAUGCCAGUCUUGGUA------------------GACUCGGUAGACCUGGUGGAAAACCCAACCAGUCCAAGAGUCG .((..(((((.((((((........)))))).)))))))......((((......))))------------------(((((....(((.((((((.....)).)))))))...))))). ( -31.40) >DroEre_CAF1 15370 111 - 1 GGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCCACAAAUGCCAGUCUUGGUA---------GAUUCGGCAGACUCGGCAGACCUGGUGGAAAGCCCAACCAGUCCGAGAGUCG (((..(((((.((((((........)))))).)))))))).....(((((((((....)---------)))).))))(((((..(.(((.((((((.....)).))))))).).))))). ( -42.00) >DroYak_CAF1 15475 120 - 1 UGCCAGUGGGUUAAGUUGCCUAAAAGGCUUAUCCUAUGCCACAAAUGCCAGUCUUGGUAGACGCGGUAGAUGCGGUAGACUCGGCAGACCUGGUGGAAAACCCAACCAGUCCAAGAGUCG .(((..(((((......)))))...)))........((((.((..((((.((((....))))..))))..)).))))(((((....(((.((((((.....)).)))))))...))))). ( -44.20) >consensus UGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCCACAAAUGCCAGUCUUGGUA__________________GACUCGGUAGACCUGGUGGAAAACCCAACCAGUCCAAGAGUCG .((..(((((.((((((........)))))).)))))))......((((......))))..................(((((....(((.((((((.....)).)))))))...))))). (-29.52 = -29.72 + 0.20)


| Location | 5,898,611 – 5,898,730 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.42 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5898611 119 - 22224390 AAAUCAUGGGAAACCCUAAUUACCGUAGC-ACAAACCAAACAGUAAGCAAACGUUUACUUUUCGUUAAUUUAGCAAUGGCUGCUAGUGGGUUAACUUGGCUAAAAGGCUUAUCCUAUGCC ....((((((((((((..((((..(((((-.((........(((((((....)))))))....((((...))))..)))))))))))))))).....((((....))))..))))))).. ( -29.90) >DroSec_CAF1 15343 119 - 1 AAAUCAUGGGAAACCCUAAUUACCGUAGC-ACAAACCAAACAGUAAGCAAACGUUUACUUUUCGCUAAUUUAGCAAUGGCUGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCC ....((((((((((((..((((..(((((-.((........(((((((....)))))))....((((...))))..)))))))))))))))).....((((....))))..))))))).. ( -32.30) >DroSim_CAF1 12650 119 - 1 AAAUCAUGGGAAACCCUAAUUACCGUAGC-ACAAACCGAACAGUAAGCAAACGUUUACUUUUCGCUAAUUUAGCAAUGGCUGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCC ....((((((((((((..((((..(((((-.((....(((.(((((((....))))))).)))((((...))))..)))))))))))))))).....((((....))))..))))))).. ( -36.00) >DroEre_CAF1 15441 119 - 1 AAAUCAUGGGAAACUCUAAUUACCGCAGC-AAAAACGAAACAGUAAGCAAACGUUUACUUUUCGCUAAUUUAGCAACGGCGGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCC ....((((((((((((..((((((((...-......((((.(((((((....)))))))))))((((...))))....)))).))))))))).....((((....))))..))))))).. ( -34.00) >DroYak_CAF1 15555 120 - 1 AAAUCAUGAGAAACUCUAAUUACCGUAGCGAAAAACAAAGCAGUGAGCAAACGUUUACUUUUCGCUAAUUUAGCAACGGUUGCCAGUGGGUUAAGUUGCCUAAAAGGCUUAUCCUAUGCC ....((((.((..........((((((((((((........(((((((....)))))))))))))).........))))).(((..(((((......)))))...)))...)).)))).. ( -28.40) >consensus AAAUCAUGGGAAACCCUAAUUACCGUAGC_ACAAACCAAACAGUAAGCAAACGUUUACUUUUCGCUAAUUUAGCAAUGGCUGCUAGUGGGUUAAGUUGGCUAAAAGGCUUAUCCUAUGCC ....((((((((((((..((((..(((((............(((((((....)))))))....((((...))))....)))))))))))))).....((((....))))..))))))).. (-25.50 = -25.42 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:57 2006