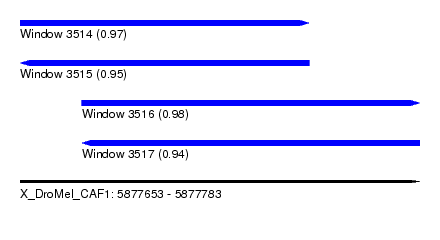
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,877,653 – 5,877,783 |
| Length | 130 |
| Max. P | 0.977643 |

| Location | 5,877,653 – 5,877,747 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.20 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
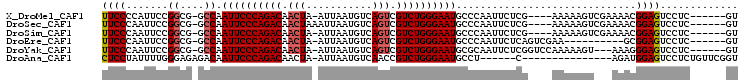
>X_DroMel_CAF1 5877653 94 + 22224390 AC------GAGGACUCCGUUUUCGACUUUUU----CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGC-CGCCGGAAUGGGGAA ..------.....((((((((((((.....)----))))))).((.(((((((((((((((((......)-)))))))))))))).)).)-)........)))).. ( -40.70) >DroSec_CAF1 82391 95 + 1 AC------GAGGACUCCGUUUUCGACUUUUU----CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAUUUAGUUGUCUGGGAAUUGGC-CGCCGGAAUUGGGAA .(------((....(((((((((((.....)----))))))..((.(((((((((((((((((.......))))))))))))))).)).)-)..)))).))).... ( -39.00) >DroSim_CAF1 85113 94 + 1 AC------GAGGACUCCGUUUUCGACUUUUU----CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGC-CGCCGGAAUUGGGAA .(------((....(((((((((((.....)----))))))..((.(((((((((((((((((......)-)))))))))))))).)).)-)..)))).))).... ( -39.40) >DroEre_CAF1 80439 88 + 1 AC------GAGGACUCCGC----------UUCGACUGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGC-CGCCGGAAUUGGGAA .(------((((......)----------)))).(..(...((.(((((((((((((((((((......)-)))))))))))))))....-.))).)).)..)... ( -35.60) >DroYak_CAF1 88699 95 + 1 AC------GAGGACUCCCUUU---ACUUUUUGGACCGAGAAUUGCGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGC-CGCCGGAAUUGGGAA ..------......((((...---.((....)).(((.(....((.(((((((((((((((((......)-)))))))))))))).))))-..))))....)))). ( -34.80) >DroAna_CAF1 87086 84 + 1 ACCGAACAGAGGACUCCAUCU---------------G------AGGCAUUCCCAGACGGUUGACAUUAAU-UAGUUGUCUGGGAAUUGUCUCUCCCAAAAUAGGAG ......((((((...)).)))---------------)------((((((((((((((..((((......)-)))..))))))))).)))))((((.......)))) ( -30.50) >consensus AC______GAGGACUCCGUUU___ACUUUUU____CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU_UAGUUGUCUGGGAAUUGGC_CGCCGGAAUUGGGAA ..............(((...............................((((((((((((((.........))))))))))))))((((....)))).....))). (-24.28 = -24.20 + -0.08)


| Location | 5,877,653 – 5,877,747 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5877653 94 - 22224390 UUCCCCAUUCCGGCG-GCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG----AAAAAGUCGAAAACGGAGUCCUC------GU ......(((((((.(-((..((((((((((.(((.-((....)).))).)))))))))))))).....(((----(.....))))...))))))....------.. ( -32.00) >DroSec_CAF1 82391 95 - 1 UUCCCAAUUCCGGCG-GCCAAUUCCCAGACAACUAAAUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG----AAAAAGUCGAAAACGGAGUCCUC------GU ......(((((((.(-((..((((((((((.(((...........))).)))))))))))))).....(((----(.....))))...))))))....------.. ( -30.80) >DroSim_CAF1 85113 94 - 1 UUCCCAAUUCCGGCG-GCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG----AAAAAGUCGAAAACGGAGUCCUC------GU ......(((((((.(-((..((((((((((.(((.-((....)).))).)))))))))))))).....(((----(.....))))...))))))....------.. ( -32.00) >DroEre_CAF1 80439 88 - 1 UUCCCAAUUCCGGCG-GCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCAGUCGAA----------GCGGAGUCCUC------GU ......(((((((.(-((..((((((((((.(((.-((....)).))).))))))))))))))..(((......)))----------.))))))....------.. ( -29.30) >DroYak_CAF1 88699 95 - 1 UUCCCAAUUCCGGCG-GCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCGCAAUUCUCGGUCCAAAAAGU---AAAGGGAGUCCUC------GU (((((....((((..-((((.(((((((((.(((.-((....)).))).))))))))))).)).....))))..(.....).---...))))).....------.. ( -27.50) >DroAna_CAF1 87086 84 - 1 CUCCUAUUUUGGGAGAGACAAUUCCCAGACAACUA-AUUAAUGUCAACCGUCUGGGAAUGCCU------C---------------AGAUGGAGUCCUCUGUUCGGU (((((.....))))).....((((((((((.....-.............))))))))))(((.------(---------------(((.((...))))))...))) ( -26.57) >consensus UUCCCAAUUCCGGCG_GCCAAUUCCCAGACAACUA_AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG____AAAAAGU___AAACGGAGUCCUC______GU ((((.......((....)).((((((((((.(((...........))).))))))))))..............................))))............. (-20.42 = -20.78 + 0.36)


| Location | 5,877,673 – 5,877,783 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -42.47 |
| Consensus MFE | -28.39 |
| Energy contribution | -30.98 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5877673 110 + 22224390 UUUUU----CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGCCGCCGGAAUGGGGAAUU-UUCCUGUUUUUGGGAAGAGGAUGCAUCC---AAUGCG (((((----(.......((.(((((((((((((((((......)-)))))))))))))).)).)).((((((..((((...-.))))..))))))))))))...((((..---.)))). ( -43.90) >DroSec_CAF1 82411 111 + 1 UUUUU----CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAUUUAGUUGUCUGGGAAUUGGCCGCCGGAAUUGGGAAUU-UUCCUGUUUUUGGGGAGAGGCUGCAUCC---AAUGCG .....----((...(((((((((((((((((((((((.......)))))))))))))))).((((.((((((..((((...-.))))..)))))).....))))))..))---))).)) ( -44.00) >DroSim_CAF1 85133 110 + 1 UUUUU----CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGCCGCCGGAAUUGGGAAUU-UUCCUGCUUUUGGGGAGAGGCUGCAUCC---AAUGCG .....----((...(((((((((((((((((((((((......)-))))))))))))))).((((.((((((..((((...-.))))..)))))).....))))))..))---))).)) ( -44.40) >DroEre_CAF1 80452 110 + 1 ---UUCGACUGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGCCGCCGGAAUUGGGAAUU-UUCCUG-UUUUGCCUAGAGGCUGCAUCC---AAUGCA ---(((......)))..((.(((((((((((((((((......)-)))))))))))))).)).))((.(((...((((...-.))))(-(...(((....))).)).)))---...)). ( -41.70) >DroYak_CAF1 88716 114 + 1 UUUUUGGACCGAGAAUUGCGCAUUCCCAGACGACUGACAUUAAU-UAGUUGUCUGGGAAUUGGCCGCCGGAAUUGGGAAUUUUUCCUG-UUUUGACUAGUGGCUGCAUCC---AAUGCA ...(((((........(((..((((((((((((((((......)-))))))))))))))).((((((((((((.((((....)))).)-)))))....))))))))))))---)).... ( -44.30) >DroPer_CAF1 105659 93 + 1 UCCGU----C-----UCCCGGCUCCCCAGGCGAUUGACAUUAAU-UAGUUGUCUGGGAAUUUGCCUCCCCAGUUGGU-------------AUUGGGAAGAGGC---AUUCGAUAAUGCU .....----.-----....(((..(((((((((((((......)-))))))))))))....((((((((((((....-------------))))))..)))))---).........))) ( -36.50) >consensus UUUUU____CGAGAAUUGGGCAUUCCCAGACGACUGACAUUAAU_UAGUUGUCUGGGAAUUGGCCGCCGGAAUUGGGAAUU_UUCCUG_UUUUGGGAAGAGGCUGCAUCC___AAUGCG .....................(((((((((((((((.........))))))))))))))).((((.((((((.(((((.....))))).)))))).....))))((((......)))). (-28.39 = -30.98 + 2.60)


| Location | 5,877,673 – 5,877,783 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -31.21 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.70 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5877673 110 - 22224390 CGCAUU---GGAUGCAUCCUCUUCCCAAAAACAGGAA-AAUUCCCCAUUCCGGCGGCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG----AAAAA .((...---(((((.......((((........))))-.......)))))..))(((..((((((((((.(((.-((....)).))).))))))))))))).........----..... ( -31.04) >DroSec_CAF1 82411 111 - 1 CGCAUU---GGAUGCAGCCUCUCCCCAAAAACAGGAA-AAUUCCCAAUUCCGGCGGCCAAUUCCCAGACAACUAAAUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG----AAAAA .(.(((---((..((.(((..(((.........))).-.....((......)).)))..((((((((((.(((...........))).))))))))))))))))).)...----..... ( -28.10) >DroSim_CAF1 85133 110 - 1 CGCAUU---GGAUGCAGCCUCUCCCCAAAAGCAGGAA-AAUUCCCAAUUCCGGCGGCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG----AAAAA .((((.---..))))...............((.((((-.........)))).))(((..((((((((((.(((.-((....)).))).))))))))))))).........----..... ( -30.20) >DroEre_CAF1 80452 110 - 1 UGCAUU---GGAUGCAGCCUCUAGGCAAAA-CAGGAA-AAUUCCCAAUUCCGGCGGCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCAGUCGAA--- .(((((---((.(((.(((....)))....-..((((-.........)))).))).))))(((((((((.(((.-((....)).))).))))))))))))................--- ( -33.90) >DroYak_CAF1 88716 114 - 1 UGCAUU---GGAUGCAGCCACUAGUCAAAA-CAGGAAAAAUUCCCAAUUCCGGCGGCCAAUUCCCAGACAACUA-AUUAAUGUCAGUCGUCUGGGAAUGCGCAAUUCUCGGUCCAAAAA ....((---((((...(((....((....)-).((((..........))))))).((((.(((((((((.(((.-((....)).))).))))))))))).))........))))))... ( -34.50) >DroPer_CAF1 105659 93 - 1 AGCAUUAUCGAAU---GCCUCUUCCCAAU-------------ACCAACUGGGGAGGCAAAUUCCCAGACAACUA-AUUAAUGUCAAUCGCCUGGGGAGCCGGGA-----G----ACGGA ............(---(((((..((((..-------------......)))))))))).....((.((((....-.....))))..((.(((((....))))).-----)----).)). ( -29.50) >consensus CGCAUU___GGAUGCAGCCUCUACCCAAAA_CAGGAA_AAUUCCCAAUUCCGGCGGCCAAUUCCCAGACAACUA_AUUAAUGUCAGUCGUCUGGGAAUGCCCAAUUCUCG____AAAAA .(((((....))))).(((.((...........(((.....))).......)).)))..((((((((((.(((...........))).))))))))))..................... (-19.60 = -20.70 + 1.10)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:48 2006