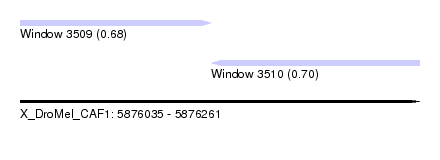
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,876,035 – 5,876,261 |
| Length | 226 |
| Max. P | 0.697745 |

| Location | 5,876,035 – 5,876,143 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -23.77 |
| Energy contribution | -25.90 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5876035 108 + 22224390 GCGCCCACAAGAUCGCCAGAUUCCUGCCAAUAAGGAAUUGCAUUUGUGCGCAGCGCACUGCGAUGGCGAAUGAGUUGGCCAA--AACCGGUUGCCGCACUUGGUUAACUU .((((((((((...((..(((((((.......))))))))).))))))(((((....)))))..))))...(((((((((((--...(((...)))...))))))))))) ( -39.10) >DroSec_CAF1 80817 108 + 1 GCGCCCACAAGAUCGCCAGAUUCCUGCCAAUAAGGAUUUGCAUUUGUGCGCAGCGCACUGCGAUGGCGAAUGAGUUGGGCAA--AACCGGUUGCCGCACUUGGUUAACUU ..((((((..(.((((((...((((.......)))).(((((...(((((...)))))))))))))))).)..).)))))..--....((((((((....))).))))). ( -36.30) >DroSim_CAF1 83507 108 + 1 GCGCCCGCAAGAUCGCCAGAUUCCUGCCAAUAAGGAUUUGCAUUUGUGCGCAGCGCACUGCGAUGGCGAAUGAGUUGGCCAA--AACCGGUUGCCGCACUUGGUUAACUU .(((((....)((((((((((.(((.......)))))))).....(((((...))))).)))))))))...(((((((((((--...(((...)))...))))))))))) ( -38.40) >DroYak_CAF1 86748 88 + 1 GCGCCCACAAGUUGGCCAGAUUCCUGCCAACAAGGAUCUGCAUUCGCGC------------CAU------GGAGUUGGCCAAAAAACCGGUUA----ACUUGGCCAGCUU ((((......((((((.((....))))))))..((((....))))))))------------...------.(((((((((((...........----..))))))))))) ( -32.12) >consensus GCGCCCACAAGAUCGCCAGAUUCCUGCCAAUAAGGAUUUGCAUUUGUGCGCAGCGCACUGCGAUGGCGAAUGAGUUGGCCAA__AACCGGUUGCCGCACUUGGUUAACUU .((((((((((...((.((((.(((.......))))))))).))))))(((((....)))))..))))...(((((((((((.....((.....))...))))))))))) (-23.77 = -25.90 + 2.13)

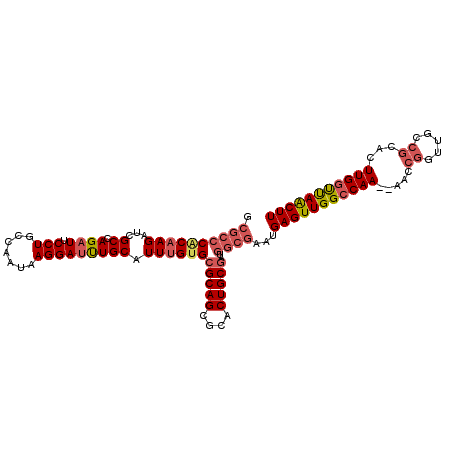
| Location | 5,876,143 – 5,876,261 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.88 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -13.76 |
| Energy contribution | -14.82 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5876143 118 - 22224390 UUUAAUGUCCAAUGUCCACUUUAAUCGUAGUGUAAAAUGAAAAGUUUAACUUGACUUAGUGACUUAGUGACUCAUGCGCGGGGUUUGUGGUUAGACUUUCGUGUGGCGCAAGCUGGAA .......(((......((((........))))....((((((.((((((((.((((..(((.....(((...))).)))..))))...))))))))))))))..(((....)))))). ( -29.40) >DroSec_CAF1 80925 93 - 1 U--------------------UAAGCGUAGUGUAAAAUGAAAAGUUUAACUUGGCUUGGGGAC---GUGACUCAUGCCCG--GUUUGUGGUUAGACUUUCGUGUGGCGCAAGCUGGAA .--------------------..(((...((((...((((((.((((((((.((((..(((.(---(((...))))))))--)))...))))))))))))))...))))..))).... ( -30.40) >DroSim_CAF1 83615 93 - 1 U--------------------UAAGCGUAGUGUAAAAUGAAAAGUUUAACUUGGCUUGGUGAC---AUGACUCAUGCCCG--GUUUGUGGUUAGACUUUCGUGUGGCGCAAGCUGGAA .--------------------..(((...((((...((((((.((((((((.((((.((.(.(---(((...))))))))--)))...))))))))))))))...))))..))).... ( -27.90) >DroYak_CAF1 86836 99 - 1 UUUAUGACUCCAUAUUUACUAGAAUUCUAGUGUAAAACUAAAAGUUUAACC----------AU---AUGACGCAUG------UUUUGUGCUUAGACUUUCGAGUGGCGCAAGCUGGAA ........(((.....((((((....))))))....(((.(((((((((.(----------((---(.(((....)------)).)))).)))))))))..)))(((....)))))). ( -26.40) >consensus U____________________UAAGCGUAGUGUAAAAUGAAAAGUUUAACUUGGCUUGGUGAC___AUGACUCAUGCCCG__GUUUGUGGUUAGACUUUCGUGUGGCGCAAGCUGGAA .............................((((...((((((.((((((((.(((...(((.....(((...))).)))...)))...))))))))))))))...))))......... (-13.76 = -14.82 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:42 2006