| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,857,302 – 5,857,403 |
| Length | 101 |
| Max. P | 0.638239 |

| Location | 5,857,302 – 5,857,403 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -25.95 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
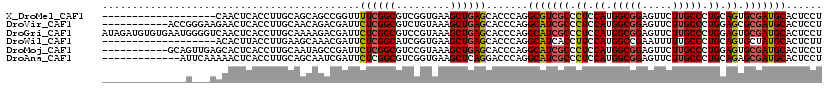
>X_DroMel_CAF1 5857302 101 + 22224390 -------------------CAACUCACCUUGCAGCAGCCGGUUUUCGGCGUCGGUGAAGCUGAGCACCCAGGCGUCGCCCUCCAUGGCGGAGUUCUUGCCCUGCAGUGCGAUGCACUCCU -------------------...((((.(((.((.(.((((.....))))...).))))).)))).......(((((((.((.((.(((((.....))))).)).)).)))))))...... ( -35.30) >DroVir_CAF1 85442 109 + 1 -----------ACCGGGAAGAACUCACCUUGCAACAGACGAUUCUCGGCGUCUGUAAAGCUGAGCACCCAGGCAUCGCCCUCCAUGGCGGAGUUCUUGCCCUGGAGCGCGAUGCACUCCU -----------...(((.....((((.(((...(((((((........))))))).))).))))..)))..(((((((.(((((.(((((.....))))).))))).)))))))...... ( -47.80) >DroGri_CAF1 84896 120 + 1 AUAGAUGUGUGAAUGGGGUCAACUCACCUUGCAAAAGACGAUUCUCGGCGUCCGUAAAGCUGAGCACCCAGGCAUCGCCCUCCAUGGCGGAGUUCUUGCCCUGGAGUGCGAUGCACUCCU ...............((((...((((.(((......((((........))))....))).)))).))))..(((((((.(((((.(((((.....))))).))))).)))))))...... ( -43.50) >DroWil_CAF1 68761 101 + 1 -------------------ACACUUACCUUGAAGCAAACGAUUCUCGGCAUCGGUGAAGCUGAGCACCCAGGCAUCACCUUCCAUGGCCGAAUUUUUGCCCUGCAGUGCUAUGCACUCUU -------------------..............(((((.(((..(((((....(.((((.(((((......)).))).)))))...))))))))))))).....(((((...)))))... ( -24.70) >DroMoj_CAF1 80341 109 + 1 -----------GCAGUUGAGCACUCACCUUGCAAUAGCCGAUUCUCGGCGUCCGUAAAGCUGAGCACCCAGGCAUCGCCCUCCAUGGCGGAGUUCUUGCCCUGGAGUGCGAUGCACUCCU -----------......(((..(((((.((((....((((.....))))....)))).).)))).......(((((((.(((((.(((((.....))))).))))).))))))).))).. ( -43.50) >DroAna_CAF1 65992 107 + 1 -------------AUUCAAAAACUCACCUUGCAGCAAUCGAUUCUCGGCGUCGGUGAAGCUCAGGACCCAGGCAUCGCCCUCCAUGGCGGAGUUCUUGCCCUGCAGAGCGAUGCACUCCU -------------..............((((.(((.((((((.......))))))...)))))))......(((((((.((.((.(((((.....))))).)).)).)))))))...... ( -33.00) >consensus ______________G___AAAACUCACCUUGCAACAGACGAUUCUCGGCGUCGGUAAAGCUGAGCACCCAGGCAUCGCCCUCCAUGGCGGAGUUCUUGCCCUGCAGUGCGAUGCACUCCU ...........................................((((((.........)))))).......(((((((.((.((.(((((.....))))).)).)).)))))))...... (-25.95 = -26.20 + 0.25)

| Location | 5,857,302 – 5,857,403 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -25.83 |
| Energy contribution | -25.55 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5857302 101 - 22224390 AGGAGUGCAUCGCACUGCAGGGCAAGAACUCCGCCAUGGAGGGCGACGCCUGGGUGCUCAGCUUCACCGACGCCGAAAACCGGCUGCUGCAAGGUGAGUUG------------------- .(.(((((...))))).).((((..(..((((.....))))..)...))))....(((((.(((((.((..((((.....)))))).)).))).)))))..------------------- ( -35.90) >DroVir_CAF1 85442 109 - 1 AGGAGUGCAUCGCGCUCCAGGGCAAGAACUCCGCCAUGGAGGGCGAUGCCUGGGUGCUCAGCUUUACAGACGCCGAGAAUCGUCUGUUGCAAGGUGAGUUCUUCCCGGU----------- ......(((((((.(((((.(((.........))).))))).)))))))(((((.(((((.(((.(((((((........)))))))...))).)))))....))))).----------- ( -54.30) >DroGri_CAF1 84896 120 - 1 AGGAGUGCAUCGCACUCCAGGGCAAGAACUCCGCCAUGGAGGGCGAUGCCUGGGUGCUCAGCUUUACGGACGCCGAGAAUCGUCUUUUGCAAGGUGAGUUGACCCCAUUCACACAUCUAU ..(((((((((((.(((((.(((.........))).))))).)))))))..(((((((((.(((...(((((........))))).....))).)))))..)))).)))).......... ( -46.00) >DroWil_CAF1 68761 101 - 1 AAGAGUGCAUAGCACUGCAGGGCAAAAAUUCGGCCAUGGAAGGUGAUGCCUGGGUGCUCAGCUUCACCGAUGCCGAGAAUCGUUUGCUUCAAGGUAAGUGU------------------- ...........(((((((.(((((((..((((((.......(((((.((.(((....))))).)))))...)))))).....)))))))....)).)))))------------------- ( -29.80) >DroMoj_CAF1 80341 109 - 1 AGGAGUGCAUCGCACUCCAGGGCAAGAACUCCGCCAUGGAGGGCGAUGCCUGGGUGCUCAGCUUUACGGACGCCGAGAAUCGGCUAUUGCAAGGUGAGUGCUCAACUGC----------- .(.((((((((((.(((((.(((.........))).))))).))))))).((((..((((.((((.(((..((((.....))))..))).))))))))..))))))).)----------- ( -50.50) >DroAna_CAF1 65992 107 - 1 AGGAGUGCAUCGCUCUGCAGGGCAAGAACUCCGCCAUGGAGGGCGAUGCCUGGGUCCUGAGCUUCACCGACGCCGAGAAUCGAUUGCUGCAAGGUGAGUUUUUGAAU------------- ((((.(((((((((((.((.(((.........))).)).))))))))))...).))))(((((.((((...(((((.......)))..))..)))))))))......------------- ( -38.80) >consensus AGGAGUGCAUCGCACUCCAGGGCAAGAACUCCGCCAUGGAGGGCGAUGCCUGGGUGCUCAGCUUCACCGACGCCGAGAAUCGUCUGCUGCAAGGUGAGUUCU___C______________ .(((((.....(((((...(((((.(..((((.....))))..)..))))).)))))...))))).....((((..................))))........................ (-25.83 = -25.55 + -0.27)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:35 2006