
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,819,196 – 5,819,329 |
| Length | 133 |
| Max. P | 0.996614 |

| Location | 5,819,196 – 5,819,290 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.17 |
| Mean single sequence MFE | -38.16 |
| Consensus MFE | -24.74 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5819196 94 + 22224390 AAUGUGCGGGCAGCCAUGAAGC---GACUUUACUGCCGUUUCAGGGAUACAGCUCA-CA-UUGCACCCAGGGAUCUGGGU---------------------CCUUGGGUGUGGCAGAUUU (((.(((((((..((.((((((---(.(......).))))))).)).....)))).-..-..((((((((((((....))---------------------)))))))))).))).))). ( -42.10) >DroSec_CAF1 24765 73 + 1 ---------------------C---GACUUUACUGCCGUCCCAGGGAUACAGCUCA-CA-UUGCACCCAGGGAUUUGGGU---------------------CCUUGGGUGUGGCAGGUUU ---------------------.---.......(((((((((...))))........-..-..((((((((((((....))---------------------))))))))))))))).... ( -32.40) >DroSim_CAF1 26674 72 + 1 ---------------------C---GACUUUACUGCCGUCCCAGGGAUACAGCUCA-CA-UUGCACCCAGGGAUCUGGGU---------------------CCUUGGGUGUGGCAGGU-U ---------------------.---.......(((((((((...))))........-..-..((((((((((((....))---------------------)))))))))))))))..-. ( -33.50) >DroEre_CAF1 24470 94 + 1 GAUGUGCGGGCAGCCGUGAACC---GACUUAACUGCCGUUCCAGGGAUACAGGGAGCCCAGGAAACCCAGGGAUCUGGGU---------------------CCUUGGGUGUGGCAG-CG- .....((((....)))).....---.......(((((..((((((((..((((...(((.((....)).))).))))..)---------------------)))))))...)))))-..- ( -38.90) >DroYak_CAF1 25314 118 + 1 GAUGUGCGGGCAGCCAUGAAGUGAAGACUCCACUGCCCUUCCAGGGAUACAGCUCA-CUGGGAAGCCCCGGGUUCUGGGUUCUGAGUUCUGAGUUCUGGCUCCUUGGGUGUGGCAGGCU- ..(.(((((((((...((.(((....))).)))))))).((((((((..(((((((-((.((((..(((((...))))))))).)))...)))..)))..))))))))....))).)..- ( -43.90) >consensus _AUGUGCGGGCAGCC_UGAA_C___GACUUUACUGCCGUUCCAGGGAUACAGCUCA_CA_UUGCACCCAGGGAUCUGGGU_____________________CCUUGGGUGUGGCAGGUUU ................................(((((..(((((((.....(.....)......((((((....)))))).....................)))))))...))))).... (-24.74 = -24.22 + -0.52)

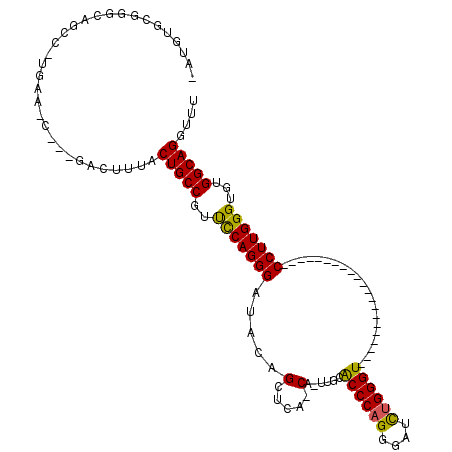
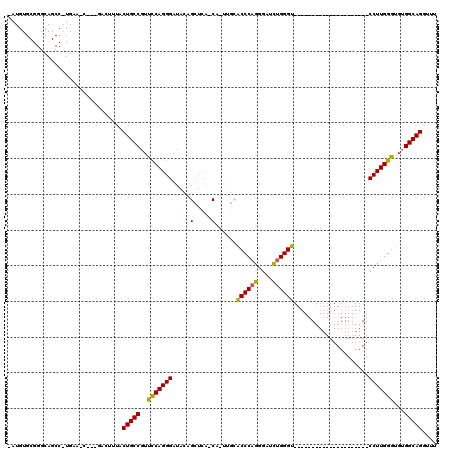
| Location | 5,819,196 – 5,819,290 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.17 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -14.88 |
| Energy contribution | -16.08 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5819196 94 - 22224390 AAAUCUGCCACACCCAAGG---------------------ACCCAGAUCCCUGGGUGCAA-UG-UGAGCUGUAUCCCUGAAACGGCAGUAAAGUC---GCUUCAUGGCUGCCCGCACAUU ..........((((((.((---------------------(......))).)))))).((-((-((....(((.((.((((.((((......)))---).)))).)).)))...)))))) ( -31.20) >DroSec_CAF1 24765 73 - 1 AAACCUGCCACACCCAAGG---------------------ACCCAAAUCCCUGGGUGCAA-UG-UGAGCUGUAUCCCUGGGACGGCAGUAAAGUC---G--------------------- ....(((((...((((.((---------------------(((((......))))((((.-..-.....))))))).))))..))))).......---.--------------------- ( -24.20) >DroSim_CAF1 26674 72 - 1 A-ACCUGCCACACCCAAGG---------------------ACCCAGAUCCCUGGGUGCAA-UG-UGAGCUGUAUCCCUGGGACGGCAGUAAAGUC---G--------------------- .-..(((((...((((.((---------------------((((((....)))))((((.-..-.....))))))).))))..))))).......---.--------------------- ( -27.10) >DroEre_CAF1 24470 94 - 1 -CG-CUGCCACACCCAAGG---------------------ACCCAGAUCCCUGGGUUUCCUGGGCUCCCUGUAUCCCUGGAACGGCAGUUAAGUC---GGUUCACGGCUGCCCGCACAUC -.(-(((((...((((.((---------------------((((((....))))))))..))))......((.((....))))))))))..((((---(.....)))))........... ( -34.30) >DroYak_CAF1 25314 118 - 1 -AGCCUGCCACACCCAAGGAGCCAGAACUCAGAACUCAGAACCCAGAACCCGGGGCUUCCCAG-UGAGCUGUAUCCCUGGAAGGGCAGUGGAGUCUUCACUUCAUGGCUGCCCGCACAUC -....(((.....(((.(((..(((..(((...(((..((((((........))).)))..))-)))))))..))).)))..((((((((((((....)))))...)))))))))).... ( -38.00) >consensus AAACCUGCCACACCCAAGG_____________________ACCCAGAUCCCUGGGUGCAA_UG_UGAGCUGUAUCCCUGGAACGGCAGUAAAGUC___G_UUCA_GGCUGCCCGCACAU_ ....(((((....(((.((.....................((((((....)))))).........((......)))))))...)))))................................ (-14.88 = -16.08 + 1.20)

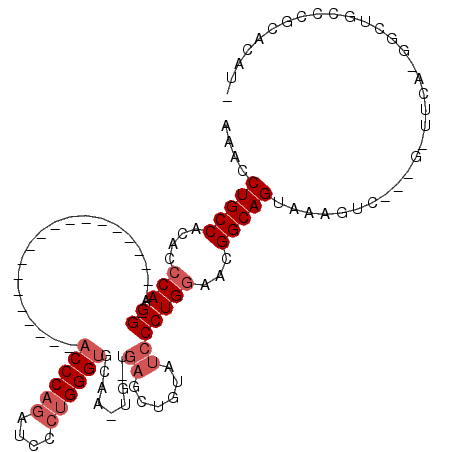
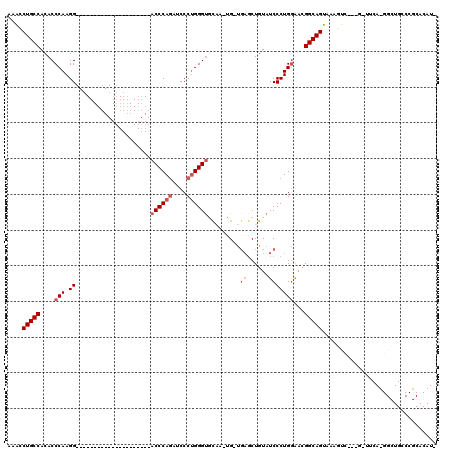
| Location | 5,819,233 – 5,819,329 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 92.41 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -34.54 |
| Energy contribution | -34.73 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5819233 96 + 22224390 UCAGGGAUACAGCUCACAUUGCACCCAGGGAUCUGGGUCCUUGGGUGUGGCAGAUUUCUGCCGG-GAAGGGCCACGAAUGCGAAACAUUGUGUGUCA .....((((((((((.....((((((((((((....))))))))))))(((((....)))))..-...))))....((((.....)))).)))))). ( -43.10) >DroSec_CAF1 24781 97 + 1 CCAGGGAUACAGCUCACAUUGCACCCAGGGAUUUGGGUCCUUGGGUGUGGCAGGUUUCUACAGGGGAACGGCCUCGAAUGCGAAACAUUGUAUGUCU ..(((.((((((.((.((((((((((((((((....))))))))))))(((..((((((....)))))).)))...)))).))....)))))).))) ( -40.10) >DroSim_CAF1 26690 95 + 1 CCAGGGAUACAGCUCACAUUGCACCCAGGGAUCUGGGUCCUUGGGUGUGGCAGGU-UCUACAGG-GAACGGCCUCGAAUGCGAAACAUUGUGUGUCU ..(((.((((((.((.((((((((((((((((....))))))))))))(((..((-(((....)-)))).)))...)))).))....)))))).))) ( -40.80) >consensus CCAGGGAUACAGCUCACAUUGCACCCAGGGAUCUGGGUCCUUGGGUGUGGCAGGUUUCUACAGG_GAACGGCCUCGAAUGCGAAACAUUGUGUGUCU ...((.((((((.((.((((((((((((((((....))))))))))))(((...((((....))))....)))...)))).))....)))))).)). (-34.54 = -34.73 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:22 2006