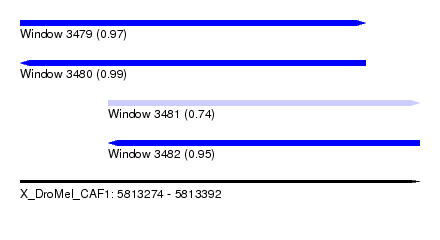
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,813,274 – 5,813,392 |
| Length | 118 |
| Max. P | 0.990911 |

| Location | 5,813,274 – 5,813,376 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5813274 102 + 22224390 UUUUCUUGCGCGGAAUGCCACAGAUUGUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGCAGUUGGUCGAUG--UGGAUGCGCU .......(((((.....(((((.....((((.....)))).((((((.((((..((((....((((...)))))))))))).))))))..))--))).))))). ( -40.20) >DroSec_CAF1 18859 102 + 1 UUUUCUUGCGCGGAAUGCCACAGAUUGUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGUUGGUGGCAGUUGGUCGAAG--UGGAUGCGCU .......(((((.....((((......((((.....)))).((((((.((((..((((....((((...)))))))))))).))))))...)--))).))))). ( -38.00) >DroSim_CAF1 19326 102 + 1 UUUUCUUGCGCGGAAUGCCACAGAUUGUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGCAGUUGGUCGAUG--UGGAUGCGCU .......(((((.....(((((.....((((.....)))).((((((.((((..((((....((((...)))))))))))).))))))..))--))).))))). ( -40.20) >DroEre_CAF1 18492 104 + 1 UUUUCUUGCGCGGAAUGCCACAGAUUGUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGAAGUUGGUCGAUGUGUGGCUGUGCU .......(((((....((((((.((((..((((((.....((((.....)))).))))))...(((((((((....)))))))))..)))).))))))))))). ( -42.80) >DroYak_CAF1 19167 102 + 1 UUUUCUUGCGCGGAAUGCCACAGAUUGUAUGUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUUGAAGUUGGUCGAUG--UGGAUGUGCU .......(((((...(((((((.......)))))))(((..(((((......)))))......(((((((........))))))))))....--....))))). ( -34.20) >consensus UUUUCUUGCGCGGAAUGCCACAGAUUGUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGCAGUUGGUCGAUG__UGGAUGCGCU .......(((((.....((((((((...)))))........((((((.((((..((((....((((...)))))))))))).))))))......))).))))). (-31.50 = -31.90 + 0.40)



| Location | 5,813,274 – 5,813,376 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -23.68 |
| Energy contribution | -24.88 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5813274 102 - 22224390 AGCGCAUCCA--CAUCGACCAACUGCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUACAAUCUGUGGCAUUCCGCGCAAGAAAA .((((.....--..........((((.....))))..((..((....((((((....)))))).((((.....)))).....))..)).....))))....... ( -31.00) >DroSec_CAF1 18859 102 - 1 AGCGCAUCCA--CUUCGACCAACUGCCACCAACAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUACAAUCUGUGGCAUUCCGCGCAAGAAAA .(((((((((--(...(.((((.((((.....(((((......)))))....)))).)))).).((((.....))))......)))).))...))))....... ( -28.30) >DroSim_CAF1 19326 102 - 1 AGCGCAUCCA--CAUCGACCAACUGCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUACAAUCUGUGGCAUUCCGCGCAAGAAAA .((((.....--..........((((.....))))..((..((....((((((....)))))).((((.....)))).....))..)).....))))....... ( -31.00) >DroEre_CAF1 18492 104 - 1 AGCACAGCCACACAUCGACCAACUUCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUACAAUCUGUGGCAUUCCGCGCAAGAAAA ......((((((.................((((....))))......((((((....)))))).((((.....)))).....))))))......(....).... ( -25.70) >DroYak_CAF1 19167 102 - 1 AGCACAUCCA--CAUCGACCAACUUCAACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCACAUACAAUCUGUGGCAUUCCGCGCAAGAAAA .((.......--..........((((........))))..((.....((((((....)))))).......(((((.......)))))......))))....... ( -21.80) >consensus AGCGCAUCCA__CAUCGACCAACUGCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUACAAUCUGUGGCAUUCCGCGCAAGAAAA .((((.................((((.....))))..((..((....((((((....)))))).((((.....)))).....))..)).....))))....... (-23.68 = -24.88 + 1.20)

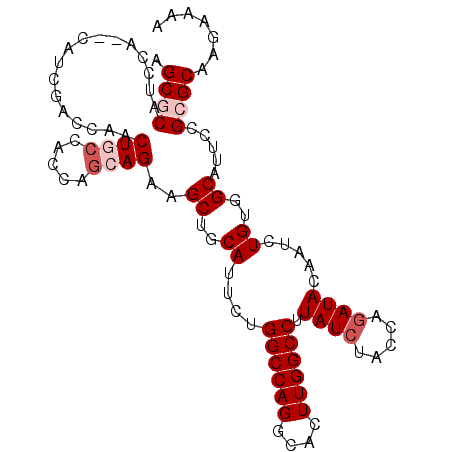

| Location | 5,813,300 – 5,813,392 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -26.48 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5813300 92 + 22224390 GUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGCAGUUGGUCGAUG--UGGAUGCGCUUACUGGGCCAGACAAU ...((((((......((((((.((((..((((....((((...)))))))))))).))))))...(--((....))).......)))))).... ( -33.40) >DroSec_CAF1 18885 92 + 1 GUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGUUGGUGGCAGUUGGUCGAAG--UGGAUGCGCUUACGGGGCCAGACAAU ...((((((......((((((.((((..((((....((((...)))))))))))).)))))).(((--((....))))).....)))))).... ( -34.40) >DroSim_CAF1 19352 92 + 1 GUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGCAGUUGGUCGAUG--UGGAUGCGCUUACUGGGCCAGACAAU ...((((((......((((((.((((..((((....((((...)))))))))))).))))))...(--((....))).......)))))).... ( -33.40) >DroEre_CAF1 18518 94 + 1 GUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGAAGUUGGUCGAUGUGUGGCUGUGCUUACUGGGCCAGACAAU ...((((((.....((((.....)))).))))))...(((((((((....))))))))).....(((.(((((..(....)..))))).))).. ( -33.80) >DroYak_CAF1 19193 92 + 1 GUAUGUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUUGAAGUUGGUCGAUG--UGGAUGUGCUUACUGGGCCAGACAAU ((((.((((.......)))).))))((((((((...(((.(.((..(..(((((......))))))--..)).))))...)).))))))..... ( -29.00) >consensus GUAUCUGGUAGAUAAGGCCAAGUGCCUGGCCAGAAUGCAGCUUCUGCUGGUGGCAGUUGGUCGAUG__UGGAUGCGCUUACUGGGCCAGACAAU ...((((((......((((((.((((..((((....((((...)))))))))))).))))))..............((....)))))))).... (-26.48 = -27.12 + 0.64)


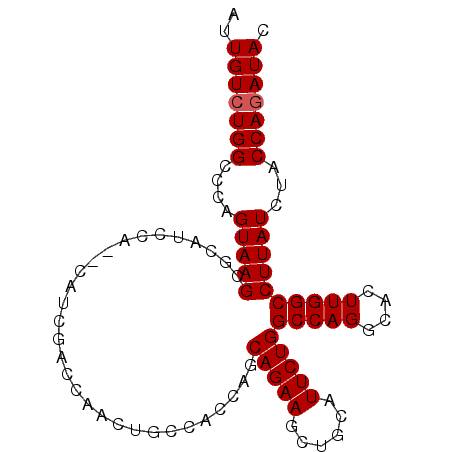
| Location | 5,813,300 – 5,813,392 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5813300 92 - 22224390 AUUGUCUGGCCCAGUAAGCGCAUCCA--CAUCGACCAACUGCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUAC ..(((((((((.((...((((.((..--....))....((((.....))))..)).))...))))))))))).((((.........)))).... ( -26.20) >DroSec_CAF1 18885 92 - 1 AUUGUCUGGCCCCGUAAGCGCAUCCA--CUUCGACCAACUGCCACCAACAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUAC ..(((((((....(((((.((.((..--....)).(((.((((.....(((((......)))))....)))).))))))))))...))))))). ( -26.10) >DroSim_CAF1 19352 92 - 1 AUUGUCUGGCCCAGUAAGCGCAUCCA--CAUCGACCAACUGCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUAC ..(((((((((.((...((((.((..--....))....((((.....))))..)).))...))))))))))).((((.........)))).... ( -26.20) >DroEre_CAF1 18518 94 - 1 AUUGUCUGGCCCAGUAAGCACAGCCACACAUCGACCAACUUCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUAC ..(((((((....................................((((....))))......((((((....)))))).......))))))). ( -22.40) >DroYak_CAF1 19193 92 - 1 AUUGUCUGGCCCAGUAAGCACAUCCA--CAUCGACCAACUUCAACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCACAUAC ..(((((((((.((...((.......--....((......))....(((....)))))...)))))))))))..(((.........)))..... ( -21.50) >consensus AUUGUCUGGCCCAGUAAGCGCAUCCA__CAUCGACCAACUGCCACCAGCAGAAGCUGCAUUCUGGCCAGGCACUUGGCCUUAUCUACCAGAUAC ..(((((((....(((((..............................(((((......)))))(((((....))))))))))...))))))). (-20.26 = -20.46 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:17 2006