
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,808,551 – 5,808,726 |
| Length | 175 |
| Max. P | 0.825814 |

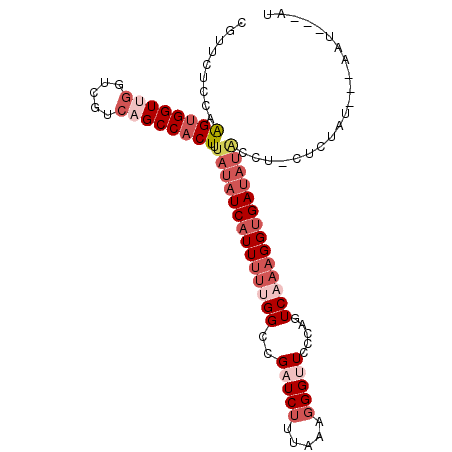
| Location | 5,808,551 – 5,808,646 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.89 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -15.93 |
| Energy contribution | -18.35 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5808551 95 - 22224390 CGUUCUCCAAGUGGUUGGUCGUCAGCCACUUUAUAUCAUUUUUGGUCGUUCUUUAAAGGGUUCCCAGUCAAAGGUGAUAUAACU-UUUAAU---AAU---UU ........(((((((((.....)))))))))((((((((((((((..(.(((.....)))....)..))))))))))))))...-......---...---.. ( -24.30) >DroPse_CAF1 41424 95 - 1 CACGCUCCAGGUGGUGGGGCUAAAGCCCCUGUACAUCCUUACCGGCCGUUCCCUGCGCGGCUCCCAUUCCAAGGUGAGUGUC--GAUCGAUGCC--CGG--- .........((((..(((((....)))))....))))....((((.((((.....((..((((((.......)).))))..)--)...)))).)--)))--- ( -30.40) >DroSec_CAF1 14072 95 - 1 CGUUCUCCAAGUGGUUGGUCGUCAGCCACUUUAUAUCAUUUUUGGCCGAUCUUUAAAGGGUUCCCAGUCAAAGGUGAUAUACCU-UUCUAU---AAU---AU ........(((((((((.....)))))))))(((((((((((((((.(((((.....)))))....)))))))))))))))...-......---...---.. ( -28.50) >DroSim_CAF1 14568 95 - 1 CGUUCUCCAAGUGGUGGGUGGUCAGCCACUUUAUAUCAUUUUUGGCCGAUCUUUAAAGGGUUCCCAGUCAAAGGUGAUAUACCU-CUCUAU---AAU---AU ..........((((.((((((....))))..(((((((((((((((.(((((.....)))))....))))))))))))))))).-..))))---...---.. ( -26.80) >DroEre_CAF1 13725 98 - 1 CGUGCUCCAAGUGGUUGGUCGUCAGCCACUUUAUAUCAUUUUUGGGCGAUCUUUAAAGGGUUCUCAGUCAAAGGUGAUAUGCAA-CUGUAA---GACAGAAU ..(((...(((((((((.....))))))))).((((((((((((((.(((((.....))))).))...))))))))))))))).-(((...---..)))... ( -28.70) >DroYak_CAF1 14383 95 - 1 CGUUCUCCAAGUGGUUGGUCGUCAGCCACUAUAUAUCAUUUUCGGGCGAUCUUUGAGGGGUUCUCAGUCAAAGGUGAUAUAAAA-CCUCAU---GAU---AU .(..((((.((((((((.....)))))))).....(((...((....))....)))))))..)...((((.((((........)-)))..)---)))---.. ( -24.60) >consensus CGUUCUCCAAGUGGUUGGUCGUCAGCCACUUUAUAUCAUUUUUGGCCGAUCUUUAAAGGGUUCCCAGUCAAAGGUGAUAUACCU_CUCUAU___AAU___AU .........((((((((.....)))))))).((((((((((((((..(((((.....))))).....))))))))))))))..................... (-15.93 = -18.35 + 2.42)

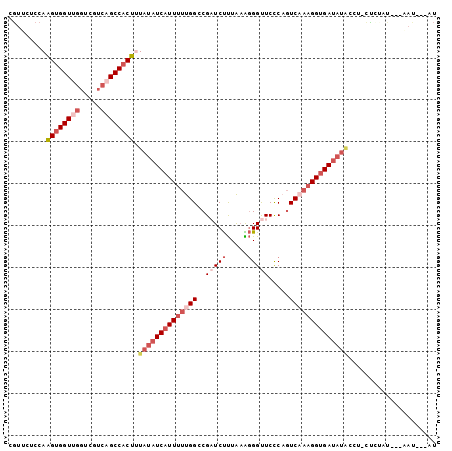
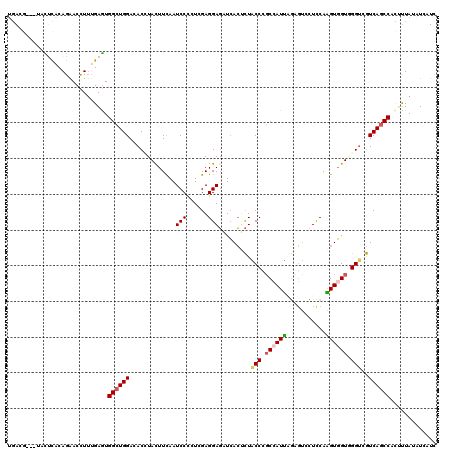
| Location | 5,808,606 – 5,808,726 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5808606 120 - 22224390 UGACGACUUCCUCACAGAACCUUUGAGUGGCUGGACACCUACUUUAAUCCACUCGAGGAGAUCACUCUACCCGCCAUUCGCGUUCUCCAAGUGGUUGGUCGUCAGCCACUUUAUAUCAUU ((((((((..(.((((((((...((((((((.((.............((.....))((((....)))).)).)))))))).)))))....))))..))))))))................ ( -35.40) >DroVir_CAF1 22017 117 - 1 UGAUC---UACCUAUAGAACCUACGAGUGGCUGGAGACGUAUUUCAAUCCCGUCGAGGAGCUCACGCUGCCCUCCAUCAAAGCAUUCCAGGUGGUAGGACGUCAGCCCUAUUACAUAAUC ...((---((....)))).((.......))(((((((((...........))))((((.((.......))))))...........)))))((((((((.(....).))))))))...... ( -29.40) >DroGri_CAF1 19973 119 - 1 UGAUGUG-UUUCCACAGAACCUUUGAGUGGCUGGAGACUUACUUCAAUCCCGUGGAGGAUUUCAACUUGCCGGCAAUUAAAUCCUUUCAGGUGGUGGGUCAUCAGCCUCACUAUUUAGUC ...((((-....))))..........(.(((((..((((((((.....((.(.(((((((((.((.(((....))))))))))))))).)).))))))))..))))).)........... ( -33.80) >DroSim_CAF1 14623 120 - 1 UGAGGAUUUCUUCACAGAACUUUUGAAUGGCUGGACACUUACUUUAAUCCCCUCGAGGAGAUCACUCUACCCGCCAUUGGCGUUCUCCAAGUGGUGGGUGGUCAGCCACUUUAUAUCAUU (((((....))))).........(((((((((((............(((.((....)).)))....((((((((((((((......)).))))))))))))))))))).))))....... ( -38.00) >DroEre_CAF1 13783 120 - 1 UGACGUCUUACUCACAGAACCUUUGAGUGGCUGGACACCUACUUCAAUCCUCUCGAGGAGAUCACUCUACCCGCCAUUCGCGUGCUCCAAGUGGUUGGUCGUCAGCCACUUUAUAUCAUU ................((......(((((((((.(((((........((((....))))(((((((....((((.....))).).....)))))))))).)))))))))))....))... ( -32.00) >DroAna_CAF1 15309 113 - 1 GUA-------CUUUCAGGACCUUCGAGUGGCUGGAAACGUACUUCAAUCCCCUGGAGGAGAUGACUCUGCCAGCCAUUAGAGUCCUUCAAGUUGUGGGUCGUCAACCCAUUUACUUUAUC ...-------.....(((((..((.(((((((((.......(((((......)))))(((....)))..))))))))).)))))))..((((.((((((.....))))))..)))).... ( -37.30) >consensus UGACG___UACUCACAGAACCUUUGAGUGGCUGGACACCUACUUCAAUCCCCUCGAGGAGAUCACUCUACCCGCCAUUAGAGUCCUCCAAGUGGUGGGUCGUCAGCCACUUUAUAUCAUC ............................((((((.............(((......))).........(((.((((((...........)))))).)))..))))))............. (-16.74 = -17.05 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:12 2006