| Sequence ID | X_DroMel_CAF1 |
|---|---|
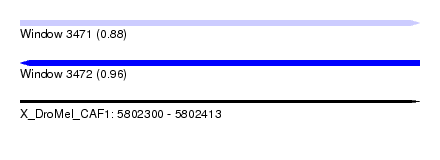
| Location | 5,802,300 – 5,802,413 |
| Length | 113 |
| Max. P | 0.961340 |

| Location | 5,802,300 – 5,802,413 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -26.70 |
| Energy contribution | -28.37 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
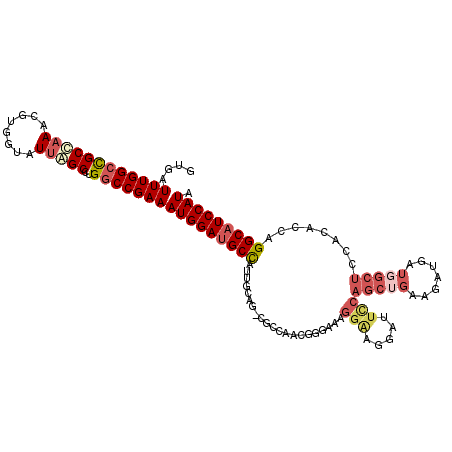
>X_DroMel_CAF1 5802300 113 + 22224390 GUGAUUUGGCCGCCAAACGUGGUAUUGGCCUGGCCGAAAUGGAUGCCAUUGCAG-CUGAAAAUGGAAAGGAAGGAUUUCAGCUGAAGAUGGUGGCUCCACACCAGGCAUCCAUA ....((((((((((((........)))))..)))))))(((((((((....(((-((((((..............)))))))))....(((((......)))))))))))))). ( -50.44) >DroSec_CAF1 7806 113 + 1 GUGAUUUGGCUGCCAAACGUGGUAUUGGCCUGGCCGAAAUGGAUGCCAUUGCGG-CUCCAAGUGGAAAGGAUGGUUUCCAGCUGGAGAUGAUGGCUCCACACCAGGCAUCCAUA ....((((((((((((........)))))..)))))))(((((((((....((.-(((((..((((((......))))))..))))).)).(((.......)))))))))))). ( -45.90) >DroSim_CAF1 8305 113 + 1 GUGAUUUGGCCGCCAAACGUGGUAUUGGCCUGGCCGAAAUGGAUGCCAUUGCGG-CUCCAAGUGGAAAGGAUGGUUUCCAGCUGGAGAUGAUGGCUCCACACCAGGCAUCCAUA ....((((((((((((........)))))..)))))))(((((((((....((.-(((((..((((((......))))))..))))).)).(((.......)))))))))))). ( -47.60) >DroEre_CAF1 7456 113 + 1 GCGAUUUGGCCGCCAAACGUGGUAUUAGCCUGGCCGAAAUGGAUGCCAUUGCAG-GGCCAACGGCACAGGGACGAUUCCAGCUGGAAAAGAUCGCUCCGCACCAGGCAUCCAUA ....(((((((((((....))))........)))))))(((((((((..(((..-.(((...)))...((..(((((((....)))....))))..)))))...))))))))). ( -43.40) >DroYak_CAF1 8123 113 + 1 GUGAUUUGGCCGCCAAACGUGGUAUUAGCCUGGCCGAAAUGGAUGCCAUUGCAG-CGCCAACGGGUAAGGAACGAUUCCAGCUGAAUAUGUUGGCUCCACACCAGGCAUCCAUA ....(((((((((((....))))........)))))))(((((((((....(((-((((....)))..(((.....))).)))).......(((.......)))))))))))). ( -39.90) >DroAna_CAF1 9036 107 + 1 GCGAUUUGGCCGCUCAGCGUGGCAUUAGCCUGUCCGAAAUGGAGGCUAUCUCCGGCGAUCACGGAUUCGGGA-------AUACGAAAAUGAGCGCCCCCUACCAGGCAUCCAUC ........(((((.....)))))....(((((........((((.....))))((((.(((....((((...-------...))))..))).))))......)))))....... ( -34.20) >consensus GUGAUUUGGCCGCCAAACGUGGUAUUAGCCUGGCCGAAAUGGAUGCCAUUGCAG_CGCCAACGGGAAAGGAAGGAUUCCAGCUGAAGAUGAUGGCUCCACACCAGGCAUCCAUA ....((((((((((((........)))))..)))))))(((((((((.....................(((.....)))(((((.......)))))........))))))))). (-26.70 = -28.37 + 1.67)

| Location | 5,802,300 – 5,802,413 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -27.42 |
| Energy contribution | -28.98 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
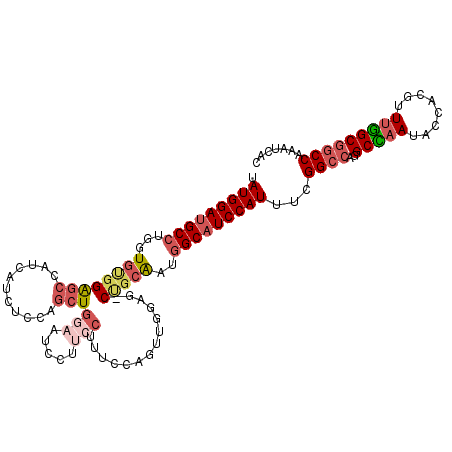
>X_DroMel_CAF1 5802300 113 - 22224390 UAUGGAUGCCUGGUGUGGAGCCACCAUCUUCAGCUGAAAUCCUUCCUUUCCAUUUUCAG-CUGCAAUGGCAUCCAUUUCGGCCAGGCCAAUACCACGUUUGGCGGCCAAAUCAC .((((((((((((((......)))))....(((((((((..............))))))-)))....)))))))))...((((..(((((........)))))))))....... ( -45.84) >DroSec_CAF1 7806 113 - 1 UAUGGAUGCCUGGUGUGGAGCCAUCAUCUCCAGCUGGAAACCAUCCUUUCCACUUGGAG-CCGCAAUGGCAUCCAUUUCGGCCAGGCCAAUACCACGUUUGGCAGCCAAAUCAC .(((((((((...(((((.........((((((.((((((......)))))).))))))-)))))..)))))))))...(((...(((((........))))).)))....... ( -47.40) >DroSim_CAF1 8305 113 - 1 UAUGGAUGCCUGGUGUGGAGCCAUCAUCUCCAGCUGGAAACCAUCCUUUCCACUUGGAG-CCGCAAUGGCAUCCAUUUCGGCCAGGCCAAUACCACGUUUGGCGGCCAAAUCAC .(((((((((...(((((.........((((((.((((((......)))))).))))))-)))))..)))))))))...((((..(((((........)))))))))....... ( -49.50) >DroEre_CAF1 7456 113 - 1 UAUGGAUGCCUGGUGCGGAGCGAUCUUUUCCAGCUGGAAUCGUCCCUGUGCCGUUGGCC-CUGCAAUGGCAUCCAUUUCGGCCAGGCUAAUACCACGUUUGGCGGCCAAAUCGC .(((((((((.((..(((..((((....(((....)))))))..)).)..))((((...-...)))))))))))))...((((..(((((........)))))))))....... ( -45.20) >DroYak_CAF1 8123 113 - 1 UAUGGAUGCCUGGUGUGGAGCCAACAUAUUCAGCUGGAAUCGUUCCUUACCCGUUGGCG-CUGCAAUGGCAUCCAUUUCGGCCAGGCUAAUACCACGUUUGGCGGCCAAAUCAC .(((((((((...((..(.((((((..........(((.....)))......)))))).-)..))..)))))))))...((((..(((((........)))))))))....... ( -44.09) >DroAna_CAF1 9036 107 - 1 GAUGGAUGCCUGGUAGGGGGCGCUCAUUUUCGUAU-------UCCCGAAUCCGUGAUCGCCGGAGAUAGCCUCCAUUUCGGACAGGCUAAUGCCACGCUGAGCGGCCAAAUCGC ..(((.(((..(((..((((((.((((.((((...-------...))))...)))).)))).....((((((((.....)))..)))))...))..)))..))).)))...... ( -35.60) >consensus UAUGGAUGCCUGGUGUGGAGCCAUCAUCUCCAGCUGGAAUCCUUCCUUUCCAGUUGGAG_CUGCAAUGGCAUCCAUUUCGGCCAGGCCAAUACCACGUUUGGCGGCCAAAUCAC .(((((((((...((((((((...........)))(((.....)))..............)))))..)))))))))...((((..(((((........)))))))))....... (-27.42 = -28.98 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:08 2006