
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,766,404 – 5,766,551 |
| Length | 147 |
| Max. P | 0.999974 |

| Location | 5,766,404 – 5,766,514 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.50 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -17.66 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5766404 110 + 22224390 ACCAAAGGAAAAAAAAUCUGAACUAAUAAAUAAGUUACCCGGGCAACAAUGCGUAUGAGUGAUAUGCAGAAAAUGCAUGUCCAUAUGCAUAUGCAUAUGCUUUCGCACAU .((...(((.......))).((((........))))....))(((...(((((((((...((((((((.....))))))))))))))))).)))...(((....)))... ( -27.00) >DroSec_CAF1 39261 109 + 1 ACCAAAGGAAUGAGUA-CAAAACGAAUAAAUAAGUUACCCGGGCAACACUGCGUAUGAGUGAUAUGCAGAAAAUGCAUGUCCAUAUGCAUAUGCACAUGCUUUCGCAUAU .((...((..((....-)).(((..........))).)).))(((....((((((((...((((((((.....))))))))))))))))..)))..((((....)))).. ( -24.50) >DroEre_CAF1 40100 96 + 1 GCCAAAGGGAAACCUAGCAAAACCAAUAAAUAAUUUAUGGAGGCAACACUGCGUAUGUGUGAUAUGCAGAAAGUGCAA------------UUGCACAUGCUA--AAAUAA (((..(((....))).......(((.(((.....)))))).)))........(((((((..((.((((.....)))))------------)..)))))))..--...... ( -29.40) >DroYak_CAF1 41870 97 + 1 AGCAGAGGAAGAAAUA-CAAAACCAAUAAAUGAUUUAGCGAGGCAACACUGCGUAUGAGUGAUAUGCAGAAAGUGCAU------------AUGCAUAUGUUUUUGCACAU .(((((((........-.....)).............(((..(((...((((((((.....))))))))....)))..------------.))).......))))).... ( -20.62) >consensus ACCAAAGGAAAAAAUA_CAAAACCAAUAAAUAAGUUACCCAGGCAACACUGCGUAUGAGUGAUAUGCAGAAAAUGCAU____________AUGCACAUGCUUUCGCACAU .........................................((((...((((((((.....))))))))...((((((((........)))))))).))))......... (-17.66 = -17.60 + -0.06)



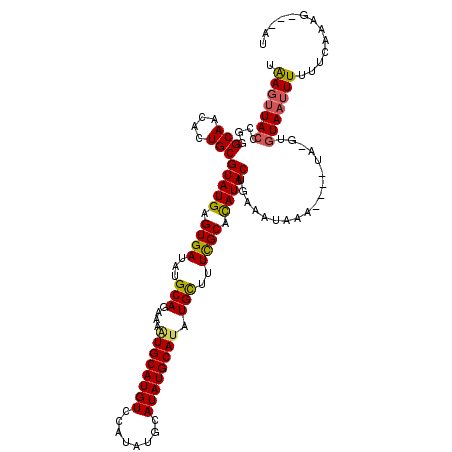
| Location | 5,766,434 – 5,766,551 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -20.89 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.73 |
| SVM decision value | 5.11 |
| SVM RNA-class probability | 0.999974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5766434 117 + 22224390 UAAGUUACCCGGGCAACAAUGCGUAUGAGUGAUAUGCAGAAAAUGCAUGUCCAUAUGCAUAUGCAUAUGCUUUCGCACAUACUGAAAUAAAUGCAUAUGUGUAAUUUUUUCAAAG---AU .(((((((...(....).(((((((((...((((((((.....)))))))))))))))))..(((((((((((((.......))))).....)))))))))))))))........---.. ( -33.50) >DroSec_CAF1 39290 113 + 1 UAAGUUACCCGGGCAACACUGCGUAUGAGUGAUAUGCAGAAAAUGCAUGUCCAUAUGCAUAUGCACAUGCUUUCGCAUAUACUAAAAUAAA----UAAGUGUAAUUUUUUCAAAG---AU .(((((((.(..(((....((((((((...((((((((.....))))))))))))))))..)))..((((....)))).............----...).)))))))........---.. ( -28.20) >DroYak_CAF1 41899 93 + 1 UGAUUUAGCGAGGCAACACUGCGUAUGAGUGAUAUGCAGAAAGUGCAU------------AUGCAUAUGUUUUUGCACAUACUGAA---------------UACUUUAUUUAAAGUGUAU ....((((...(....).((((((((.....))))))))...(((((.------------..((....))...)))))...))))(---------------((((((....))))))).. ( -23.80) >consensus UAAGUUACCCGGGCAACACUGCGUAUGAGUGAUAUGCAGAAAAUGCAUGUCCAUAUGCAUAUGCAUAUGCUUUCGCACAUACUGAAAUAAA____UA_GUGUAAUUUUUUCAAAG___AU .(((((((....(((....)))(((((.((((...(((....((((((((........)))))))).)))..)))).)))))..................)))))))............. (-20.89 = -21.40 + 0.51)


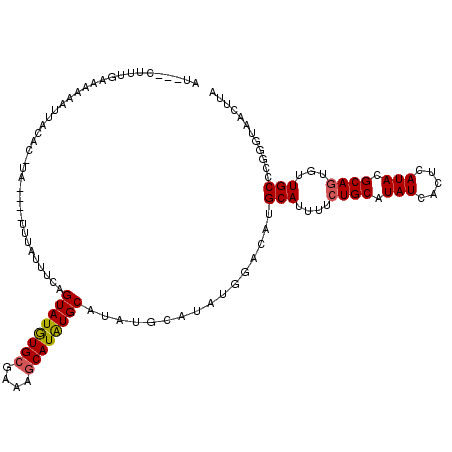
| Location | 5,766,434 – 5,766,551 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -17.37 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.64 |
| SVM decision value | 5.11 |
| SVM RNA-class probability | 0.999974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5766434 117 - 22224390 AU---CUUUGAAAAAAUUACACAUAUGCAUUUAUUUCAGUAUGUGCGAAAGCAUAUGCAUAUGCAUAUGGACAUGCAUUUUCUGCAUAUCACUCAUACGCAUUGUUGCCCGGGUAACUUA ..---...(((..........(((((((((........((((((((....))))))))..)))))))))((.(((((.....))))).))..)))........(((((....)))))... ( -34.20) >DroSec_CAF1 39290 113 - 1 AU---CUUUGAAAAAAUUACACUUA----UUUAUUUUAGUAUAUGCGAAAGCAUGUGCAUAUGCAUAUGGACAUGCAUUUUCUGCAUAUCACUCAUACGCAGUGUUGCCCGGGUAACUUA ..---....................----.........((((((((....))))))))...(((.((((((.(((((.....))))).))...)))).)))..(((((....)))))... ( -29.50) >DroYak_CAF1 41899 93 - 1 AUACACUUUAAAUAAAGUA---------------UUCAGUAUGUGCAAAAACAUAUGCAU------------AUGCACUUUCUGCAUAUCACUCAUACGCAGUGUUGCCUCGCUAAAUCA ....(((((....))))).---------------....(((((((((........)))))------------)))).......(((...(((((....).)))).)))............ ( -17.90) >consensus AU___CUUUGAAAAAAUUACAC_UA____UUUAUUUCAGUAUGUGCGAAAGCAUAUGCAUAUGCAUAUGGACAUGCAUUUUCUGCAUAUCACUCAUACGCAGUGUUGCCCGGGUAACUUA ......................................((((((((....))))))))................(((....((((.(((.....))).))))...)))............ (-17.37 = -17.60 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:53 2006