
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,764,872 – 5,765,001 |
| Length | 129 |
| Max. P | 0.793928 |

| Location | 5,764,872 – 5,764,970 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -18.66 |
| Consensus MFE | -5.14 |
| Energy contribution | -6.02 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.793928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
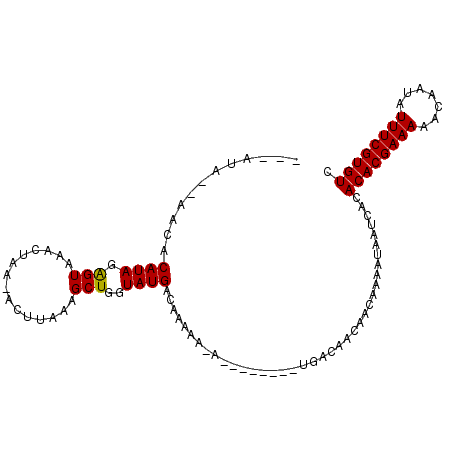
>X_DroMel_CAF1 5764872 98 - 22224390 UGGUAUGACAAAAA-A--------UGACAACAACAAAAUAAUCACACACGAAAA------ACAAUAUUUCGUGUCAAAAUUUGUGUCGUUUUUUGCUUUAUUUCCUACUGCCG .((.((((((((((-(--------((((....(((((........((((((((.------......)))))))).....))))))))))))))))..))))..))........ ( -20.42) >DroSec_CAF1 37750 97 - 1 UGGUAUGACAAAAA-A--------UGGCAAUAACAAAAUAAUCACACACGAAAA------ACAAUAUUUCGUGUCAAAAUUUGUGUCGUUU-UUGCUUUAUUUCCCACUGCCG .((((((..(((..-.--------.(((((.(((...((((....((((((((.------......))))))))......))))...))).-)))))...)))..)).)))). ( -19.60) >DroEre_CAF1 38249 106 - 1 UGGUAUGAAAAAAA-AAAAAAAACCGACAACAACAAAAUAAUCACACACGAAAA------ACAAUAUUUCGUGUCAAAAUUUGUGUCGUUUUUUGCUUCAUUUCCUACUGCUG .((((.((((..((-..(((((((.(((....(((((........((((((((.------......)))))))).....)))))))))))))))..))..)))).)))).... ( -19.82) >DroYak_CAF1 40176 107 - 1 UGGUAUGACAACAACAACAAAUACAAAUGGCAACAUUAUAAUCACACACGAAAA------ACAAUAUUUCGUGUCAAAAUUUGUGUCGUUUUUUGCUUCAUUUCCUACUGCCG .((((.((((.(((...........((((....))))........((((((((.------......))))))))......)))))))((.....))............)))). ( -20.00) >DroAna_CAF1 41509 96 - 1 UU-------AAAAG-G--------UAAAAACAAGCAAAUAAACAGCUCCGAAAAAAAAACACAAUAU-UCGUGUCAAAAUGUGUGUCGUUUUUUGCUUUAUUUCCUACUACUG ..-------...((-(--------.(((...(((((((.((((.((..((........((((.....-..)))).......)).)).)))))))))))..))))))....... ( -13.46) >consensus UGGUAUGACAAAAA_A________UGACAACAACAAAAUAAUCACACACGAAAA______ACAAUAUUUCGUGUCAAAAUUUGUGUCGUUUUUUGCUUUAUUUCCUACUGCCG .((((........................................((((((((.............))))))))........(((..((.....))..))).......)))). ( -5.14 = -6.02 + 0.88)



| Location | 5,764,910 – 5,765,001 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.52 |
| Mean single sequence MFE | -10.31 |
| Consensus MFE | -8.18 |
| Energy contribution | -7.96 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5764910 91 - 22224390 ---AUA--AACACAUAGAGUAAACUAA-ACUUAAAGCUGGUAUGACAAAAA-A--------UGACAACAACAAAAUAAUCACACACGAAAAACAAUAUUUCGUGUC ---...--....(((.((((.......-)))).....((......))....-)--------))...................((((((((.......)))))))). ( -8.30) >DroSec_CAF1 37787 92 - 1 ---GUA--AACACAUAGGGUAAACUAAAACUUAAAGCUGGUAUGACAAAAA-A--------UGGCAAUAACAAAAUAAUCACACACGAAAAACAAUAUUUCGUGUC ---...--......(((......)))...........((.(((........-)--------)).))................((((((((.......)))))))). ( -9.80) >DroYak_CAF1 40214 105 - 1 AAAAAAUCAACACAUAGAGUAAACUAA-ACUUAAAGCUGGUAUGACAACAACAACAAAUACAAAUGGCAACAUUAUAAUCACACACGAAAAACAAUAUUUCGUGUC ................((((.......-)))).......((((..............)))).((((....))))........((((((((.......)))))))). ( -12.84) >consensus ___AUA__AACACAUAGAGUAAACUAA_ACUUAAAGCUGGUAUGACAAAAA_A________UGACAACAACAAAAUAAUCACACACGAAAAACAAUAUUUCGUGUC ............((((.(((...............)))..))))......................................((((((((.......)))))))). ( -8.18 = -7.96 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:50 2006