
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,751,291 – 5,751,476 |
| Length | 185 |
| Max. P | 0.960917 |

| Location | 5,751,291 – 5,751,396 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -25.16 |
| Energy contribution | -27.60 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
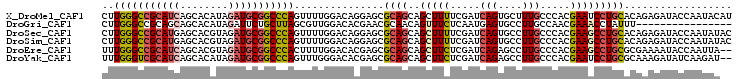
>X_DroMel_CAF1 5751291 105 + 22224390 AUGUAUUGGUAUCUCUGUGCAGGAUUCGUGGGCAAAGCACUGAUCGAAAAGCUGCUGCGCUCCUGUCCAAAACUGGGCCGCAUCUAUGUGCUGAUGCGGCCCAAG .(((((.((....)).)))))((((..(.((((..((((((........)).))))..))))).)))).....(((((((((((........))))))))))).. ( -41.90) >DroGri_CAF1 29291 89 + 1 ----------------AAAUAGGUUUCGUUGGCAAGGCACUCAUUGAGAAACUGUUGCGUUCGUGUCCAACGCUAAGCAGAAUCUAUGUGCUGCUGCGGCCCAAG ----------------.....((..((((.((((..((((......(((..((((((((((.......)))))..)))))..)))..)))))))))))).))... ( -22.80) >DroSec_CAF1 24241 105 + 1 GUAUAUUGGUAUCUCUGUGCAGGCUUCGUGGGCAAGGCACUGAUCGAAAAGCUGCUGCGCUCCUGUCCAAAACUGGGCCGCAUCUACGUGCUCAUGCGGCCCAAG .....((((.......((((((((((..((..((......))..))..))))..))))))......))))...((((((((((..........)))))))))).. ( -36.92) >DroSim_CAF1 24830 105 + 1 GUAUAUUGGUAUCUCUGUGCAGGCUUCGUGGGCAAGGCACUGAUCGAAAAGCUGCUGCGCUCCUGUCCAAAACUGGGCCGCAUCUACGUGCUCAUGCGGCCCAAG .....((((.......((((((((((..((..((......))..))..))))..))))))......))))...((((((((((..........)))))))))).. ( -36.92) >DroEre_CAF1 24743 103 + 1 --UAAUUGGUAUUUUCGCGCAGGCUUCGUGGGCAAGGCUCUGAUCGAGAAGCUGCUGCGCUCGUGUCCAAAAGUGGGCCGCAUCUACGUGCUGAUGCGGCCCAAA --...((((.((....(((((((((((.((..((......))..)).)))))..))))))....))))))...(((((((((((........))))))))))).. ( -46.10) >DroYak_CAF1 26071 103 + 1 --AUCUUGAUAUCUUUGCGCAGGAUUCGUGGGCAAGGCUCUGAUCGAGAAGCUGCUGCGCUCGUGUCCCAAACUGGGCCGCAUCUAUGUGCUGAUGCGACCCAAA --.....(((((....((((((.(((((.((((...))))....))))....).))))))..)))))......((((.((((((........)))))).)))).. ( -35.10) >consensus __AUAUUGGUAUCUCUGUGCAGGCUUCGUGGGCAAGGCACUGAUCGAAAAGCUGCUGCGCUCCUGUCCAAAACUGGGCCGCAUCUACGUGCUGAUGCGGCCCAAG .....((((.......((((((((((..((..((......))..))..))))..))))))......))))...(((((((((((........))))))))))).. (-25.16 = -27.60 + 2.45)

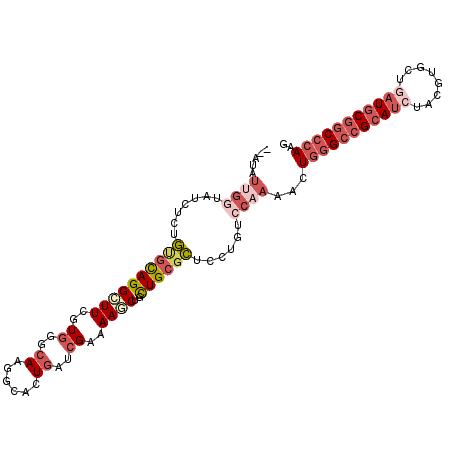
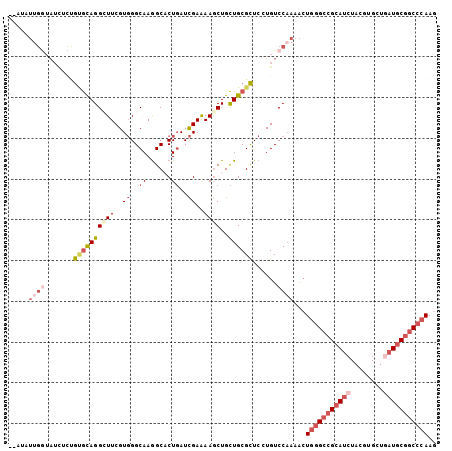
| Location | 5,751,291 – 5,751,396 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -22.77 |
| Energy contribution | -23.47 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5751291 105 - 22224390 CUUGGGCCGCAUCAGCACAUAGAUGCGGCCCAGUUUUGGACAGGAGCGCAGCAGCUUUUCGAUCAGUGCUUUGCCCACGAAUCCUGCACAGAGAUACCAAUACAU ..(((((((((((........)))))))))))((.((((...((.(((.((((.((........)))))).)))))....(((((....)).))).)))).)).. ( -40.70) >DroGri_CAF1 29291 89 - 1 CUUGGGCCGCAGCAGCACAUAGAUUCUGCUUAGCGUUGGACACGAACGCAACAGUUUCUCAAUGAGUGCCUUGCCAACGAAACCUAUUU---------------- ..((((.((..(((((((..(((..(((.((.(((((.(...).))))))))))..)))......))))..)))...))...))))...---------------- ( -19.50) >DroSec_CAF1 24241 105 - 1 CUUGGGCCGCAUGAGCACGUAGAUGCGGCCCAGUUUUGGACAGGAGCGCAGCAGCUUUUCGAUCAGUGCCUUGCCCACGAAGCCUGCACAGAGAUACCAAUAUAC .(((((((((((..........)))))))))))(((((..((((..((..(((((............))..)))...))...))))..)))))............ ( -33.70) >DroSim_CAF1 24830 105 - 1 CUUGGGCCGCAUGAGCACGUAGAUGCGGCCCAGUUUUGGACAGGAGCGCAGCAGCUUUUCGAUCAGUGCCUUGCCCACGAAGCCUGCACAGAGAUACCAAUAUAC .(((((((((((..........)))))))))))(((((..((((..((..(((((............))..)))...))...))))..)))))............ ( -33.70) >DroEre_CAF1 24743 103 - 1 UUUGGGCCGCAUCAGCACGUAGAUGCGGCCCACUUUUGGACACGAGCGCAGCAGCUUCUCGAUCAGAGCCUUGCCCACGAAGCCUGCGCGAAAAUACCAAUUA-- ..(((((((((((........)))))))))))...((((......((((((..(((((.....(((....))).....))))))))))).......))))...-- ( -41.82) >DroYak_CAF1 26071 103 - 1 UUUGGGUCGCAUCAGCACAUAGAUGCGGCCCAGUUUGGGACACGAGCGCAGCAGCUUCUCGAUCAGAGCCUUGCCCACGAAUCCUGCGCAAAGAUAUCAAGAU-- ..(((((((((((........)))))))))))((.(((((..((.(.((((..((((........)))).)))).).))..))))).))..............-- ( -34.80) >consensus CUUGGGCCGCAUCAGCACAUAGAUGCGGCCCAGUUUUGGACACGAGCGCAGCAGCUUCUCGAUCAGUGCCUUGCCCACGAAGCCUGCACAGAGAUACCAAUAU__ ..(((((((((((........)))))))))))...............((((..(((((.....(((....))).....))))))))).................. (-22.77 = -23.47 + 0.70)

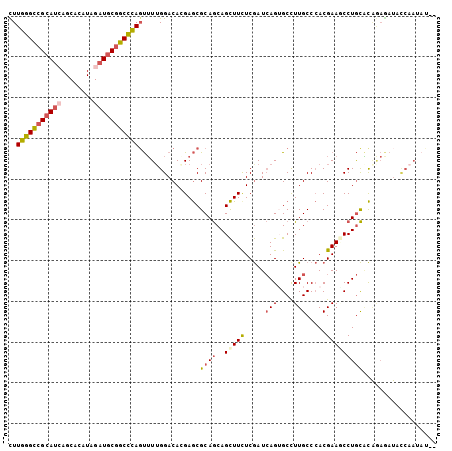
| Location | 5,751,356 – 5,751,476 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -48.15 |
| Consensus MFE | -31.35 |
| Energy contribution | -32.88 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5751356 120 + 22224390 UCCAAAACUGGGCCGCAUCUAUGUGCUGAUGCGGCCCAAGAAGGGCCACACAAUUGAGGAGCGCCUGCAGCUGCAGUGGGAAUCGAGGUUGUAUGAACGGCUGCGCAGAGAACAGCCCAA ........(((((((((((........)))))))))))....((((..........(((....)))(((((((.....(.(((....))).).....)))))))..........)))).. ( -46.50) >DroVir_CAF1 3710 120 + 1 CUACACACUGGGCAAGAUCUAUGUGCUGAUACGGCCCAAGAAGGGCGUUGCCAUCGAGCAGCGCCUGGAGGCGUUGCUCAAUUGCAAGCUGUACGAGCGAUUGCGGCGCGAGCAGCCCCA ........(((((...(((........)))...)))))....(((.(((((....((((((((((....))))))))))..((((..((((((........)))))))))))))))))). ( -54.10) >DroSim_CAF1 24895 120 + 1 UCCAAAACUGGGCCGCAUCUACGUGCUCAUGCGGCCCAAGAAGGGCCACACAAUUGAGGAGCGCCUGCAGCUGCAGUGGGAAACGAGGCUGUAUGAACGGCUGCGCAGGGAACAGCCCGA ........((((((((((..........))))))))))....((((..............((((((((....))))..(....)..((((((....)))))))))).(....).)))).. ( -46.60) >DroEre_CAF1 24806 120 + 1 UCCAAAAGUGGGCCGCAUCUACGUGCUGAUGCGGCCCAAAAAGGGCCAAACCAUUGAGGAGCGCCUGCAGCUGCAGUGGGAAACGAGGCUGUAUGAGCGGCUGCGAAGUGAGCAGCCCGA (((.....(((((((((((........)))))))))))....((......)).....)))((.(.(((((((...((.....))..))))))).).))((((((.......))))))... ( -55.60) >DroYak_CAF1 26134 120 + 1 UCCCAAACUGGGCCGCAUCUAUGUGCUGAUGCGACCCAAAAAGGGCCACACCAUUGAGGAGCGCCUGCAGCAGCAGUGGGAAACGAGGCUGUACGAACGGCUGCGACGGGAGCAGCCCGA ........((((.((((((........)))))).))))....((((....((.....)).((.((((..(((((.((.(....)..(......)..)).)))))..)))).)).)))).. ( -47.30) >DroAna_CAF1 26320 120 + 1 UCCAAAACUAAGCCGGAUUUAUGUCCUGAUGAGGCCCAAGAAGGGCAUUGAUAUCGAGGAUCGGUUGCAGCAGCAAUGGGAGACCAAGUUAUACGAUAGACUGCGUCAGGAACAGCCCGA .............(((.....((((((((((((((((.....)))).....(((((.......(((((....)))))((....))........)))))..)).))))))).)))..))). ( -38.80) >consensus UCCAAAACUGGGCCGCAUCUAUGUGCUGAUGCGGCCCAAGAAGGGCCACACCAUUGAGGAGCGCCUGCAGCUGCAGUGGGAAACGAGGCUGUACGAACGGCUGCGAAGGGAACAGCCCGA ........(((((((((((........)))))))))))....((((...............((.((((....)))))).....((.((((((....)))))).)).........)))).. (-31.35 = -32.88 + 1.53)


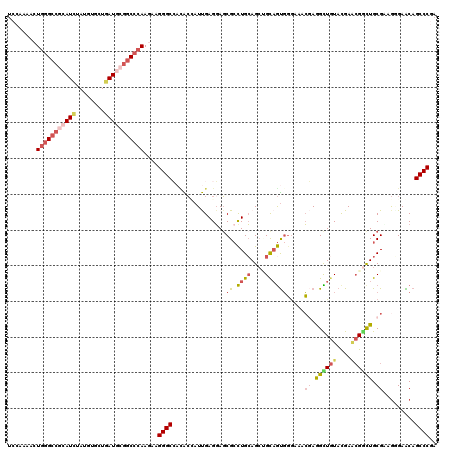
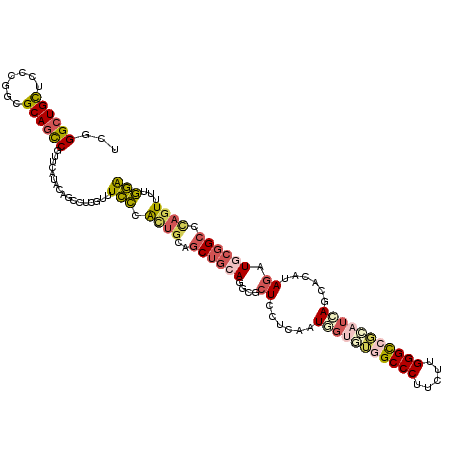
| Location | 5,751,356 – 5,751,476 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -45.24 |
| Consensus MFE | -30.97 |
| Energy contribution | -32.08 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5751356 120 - 22224390 UUGGGCUGUUCUCUGCGCAGCCGUUCAUACAACCUCGAUUCCCACUGCAGCUGCAGGCGCUCCUCAAUUGUGUGGCCCUUCUUGGGCCGCAUCAGCACAUAGAUGCGGCCCAGUUUUGGA ...((((((.......))))))..((((((((............((((....))))...........)))))))).((...((((((((((((........))))))))))))....)). ( -46.30) >DroVir_CAF1 3710 120 - 1 UGGGGCUGCUCGCGCCGCAAUCGCUCGUACAGCUUGCAAUUGAGCAACGCCUCCAGGCGCUGCUCGAUGGCAACGCCCUUCUUGGGCCGUAUCAGCACAUAGAUCUUGCCCAGUGUGUAG ((((((.((....)).))....(((.((((.(.((((.((((((((.((((....)))).)))))))).)))))((((.....)))).)))).)))............))))........ ( -48.90) >DroSim_CAF1 24895 120 - 1 UCGGGCUGUUCCCUGCGCAGCCGUUCAUACAGCCUCGUUUCCCACUGCAGCUGCAGGCGCUCCUCAAUUGUGUGGCCCUUCUUGGGCCGCAUGAGCACGUAGAUGCGGCCCAGUUUUGGA ..(((((((...((((((.((.((....)).))...........((((....))))))((((.......(((((((((.....)))))))))))))..))))..)))))))......... ( -50.11) >DroEre_CAF1 24806 120 - 1 UCGGGCUGCUCACUUCGCAGCCGCUCAUACAGCCUCGUUUCCCACUGCAGCUGCAGGCGCUCCUCAAUGGUUUGGCCCUUUUUGGGCCGCAUCAGCACGUAGAUGCGGCCCACUUUUGGA ...((((((.......)))))).........(((..........((((....)))).....((.....))...)))((....(((((((((((........))))))))))).....)). ( -46.30) >DroYak_CAF1 26134 120 - 1 UCGGGCUGCUCCCGUCGCAGCCGUUCGUACAGCCUCGUUUCCCACUGCUGCUGCAGGCGCUCCUCAAUGGUGUGGCCCUUUUUGGGUCGCAUCAGCACAUAGAUGCGGCCCAGUUUGGGA ...((((((.......))))))((..(((((((.............)))).)))..))((.((.....)).))..(((...((((((((((((........))))))))))))...))). ( -50.12) >DroAna_CAF1 26320 120 - 1 UCGGGCUGUUCCUGACGCAGUCUAUCGUAUAACUUGGUCUCCCAUUGCUGCUGCAACCGAUCCUCGAUAUCAAUGCCCUUCUUGGGCCUCAUCAGGACAUAAAUCCGGCUUAGUUUUGGA ..(((((((.......))))))).(((...((((.((((.....((((....))))..(.((((.(((......((((.....))))...))))))))........)))).)))).))). ( -29.70) >consensus UCGGGCUGCUCCCGGCGCAGCCGUUCAUACAGCCUCGUUUCCCACUGCAGCUGCAGGCGCUCCUCAAUGGUGUGGCCCUUCUUGGGCCGCAUCAGCACAUAGAUGCGGCCCAGUUUUGGA ...((((((.......)))))).................(((.((((..((((((....((......(((((((((((.....)))))))))))......)).)))))).))))...))) (-30.97 = -32.08 + 1.12)

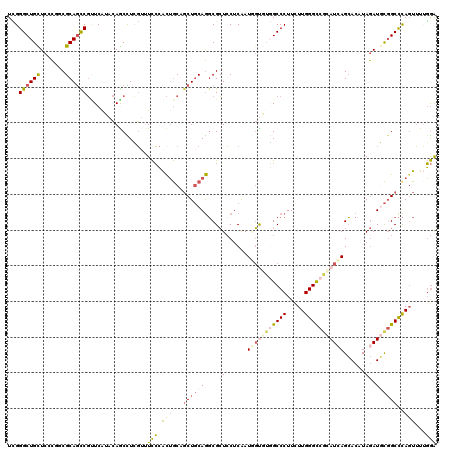
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:48 2006