| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,734,737 – 5,734,848 |
| Length | 111 |
| Max. P | 0.865724 |

| Location | 5,734,737 – 5,734,848 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -16.18 |
| Energy contribution | -17.10 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
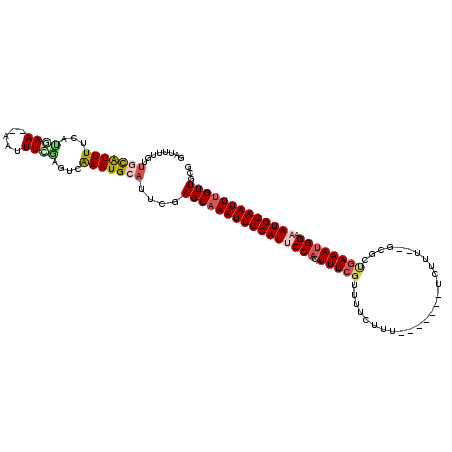
>X_DroMel_CAF1 5734737 111 + 22224390 GAUUUUGUUGCAUUUUCAUUAAAAAAUUUCGAGUCAAAUGCAUUCGAGCAAAUUGCAUUCCACUUUUCGUUUUCUUU-------UCUUU--GCGCUGAAAUGGAAAUGCAAUUUGUUGCG .........(((................(((((((....).))))))(((((((((((((((.(((.(((.......-------.....--)))..))).))).))))))))))))))). ( -26.70) >DroSim_CAF1 8472 110 + 1 GAUUUUGUUGCAUUUUCAUUAAA-AAUUUCGAGUCAAAUGCAUUCGAGCAAAUUGCAUUCCACAUUUCGUUUUCUUU-------UCUUU--GCGCUGAAAUGGAAAUGCAAUUUGUUGCG .........(((...........-....(((((((....).))))))((((((((((((.(.(((((((........-------.....--....)))))))).))))))))))))))). ( -25.73) >DroEre_CAF1 8217 109 + 1 GAUUUGGUUGCGUUUUCACGAA--AUUUUUGAGUCAAAUGCAUUCGAGCAAAUUGCAUUCCAAAUUUCGUUUUCUUU-------UCUCA--GCGCUGAAAUGGAAAUGCAAUUUGUUGCG (((..(((((((((((((((((--((((..(((((((.(((......)))..))).)))).))))))))).....((-------((...--.....)))).)))))))))))).)))... ( -26.20) >DroYak_CAF1 9168 109 + 1 GAUUUUGUUGCAUUUUGAUGAA--AUUUUCGAGUUAAAUGCAUUCGAGCAAAUUGCAUUCCCCAUUUCGUUUUCUUU-------UCUUU--GCGCCGAAAUGGAAAUGCAAUUUGUUGCG ........(((((((...(((.--....)))....)))))))..(((((((((((((((..((((((((........-------.....--....)))))))).))))))))))))).)) ( -30.13) >DroAna_CAF1 7219 108 + 1 GAUUUUGUUUUAUUUUCAUGAA----UUUUAAGAUAAAUGCAUUCGAGCAAAUUGCAU-CCACUUUUCGUUUUCUUU-------UUUGUAAAUGCUGAAAUGGAAAUGCAAUUUGUUGCG ...(((((((((..........----...))))))))).(((.....(((((((((((-(((.(((.(((((.(...-------...).)))))..))).))))..))))))))))))). ( -23.62) >DroPer_CAF1 10274 118 + 1 AAUUUUUUCUUGUUUUUGAAAAACAGUUUUAAGAUGAAUGCAUUCAAGCAAAUUGCAUACCACUUUUGGUUUUUCUUUUGGAUUUUUUU--CCCUAGAAAUGGAAAUGCAAUUCGUUGCG .((((..(((((...(((.....)))...))))).))))(((.....((.((((((((((((....)))).(((((...(((......)--))..))))).....)))))))).))))). ( -22.50) >consensus GAUUUUGUUGCAUUUUCAUGAA__AAUUUCGAGUCAAAUGCAUUCGAGCAAAUUGCAUUCCACAUUUCGUUUUCUUU_______UCUUU__GCGCUGAAAUGGAAAUGCAAUUUGUUGCG ........(((((((...((((.....))))....)))))))....((((((((((((((((..(((((..........................)))))))).)))))))))))))... (-16.18 = -17.10 + 0.92)


| Location | 5,734,737 – 5,734,848 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.89 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
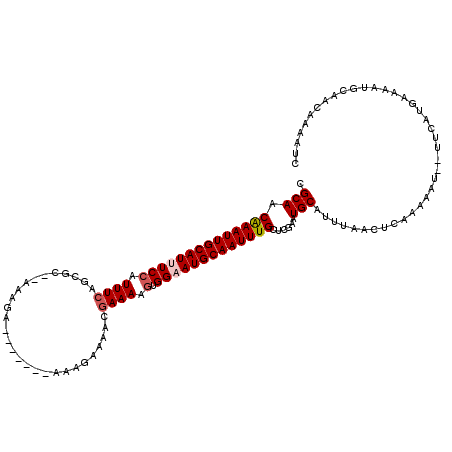
>X_DroMel_CAF1 5734737 111 - 22224390 CGCAACAAAUUGCAUUUCCAUUUCAGCGC--AAAGA-------AAAGAAAACGAAAAGUGGAAUGCAAUUUGCUCGAAUGCAUUUGACUCGAAAUUUUUUAAUGAAAAUGCAACAAAAUC ((...(((((((((((.((((((...((.--.....-------........))..)))))))))))))))))..))..(((((((....................)))))))........ ( -23.59) >DroSim_CAF1 8472 110 - 1 CGCAACAAAUUGCAUUUCCAUUUCAGCGC--AAAGA-------AAAGAAAACGAAAUGUGGAAUGCAAUUUGCUCGAAUGCAUUUGACUCGAAAUU-UUUAAUGAAAAUGCAACAAAAUC ((...(((((((((((((((((((.....--.....-------.........)))))).)))))))))))))..))..(((((((...........-........)))))))........ ( -26.02) >DroEre_CAF1 8217 109 - 1 CGCAACAAAUUGCAUUUCCAUUUCAGCGC--UGAGA-------AAAGAAAACGAAAUUUGGAAUGCAAUUUGCUCGAAUGCAUUUGACUCAAAAAU--UUCGUGAAAACGCAACCAAAUC .((..(((((((((((((.(((((....(--(....-------..)).....)))))..)))))))))))))(.((((....((((...))))...--)))).).....))......... ( -22.70) >DroYak_CAF1 9168 109 - 1 CGCAACAAAUUGCAUUUCCAUUUCGGCGC--AAAGA-------AAAGAAAACGAAAUGGGGAAUGCAAUUUGCUCGAAUGCAUUUAACUCGAAAAU--UUCAUCAAAAUGCAACAAAAUC .(((.((((((((((((((((((((...(--.....-------...)....)))))).)))))))))))))).((((...........))))....--..........)))......... ( -28.90) >DroAna_CAF1 7219 108 - 1 CGCAACAAAUUGCAUUUCCAUUUCAGCAUUUACAAA-------AAAGAAAACGAAAAGUGG-AUGCAAUUUGCUCGAAUGCAUUUAUCUUAAAA----UUCAUGAAAAUAAAACAAAAUC .(((.(((((((((..(((((((...(.(((.(...-------...).))).)..))))))-))))))))))......))).............----...................... ( -16.30) >DroPer_CAF1 10274 118 - 1 CGCAACGAAUUGCAUUUCCAUUUCUAGGG--AAAAAAAUCCAAAAGAAAAACCAAAAGUGGUAUGCAAUUUGCUUGAAUGCAUUCAUCUUAAAACUGUUUUUCAAAAACAAGAAAAAAUU ..(((((((((((((.....(((((..((--(......)))...))))).((((....)))))))))))))).))).....................((((((........))))))... ( -23.00) >consensus CGCAACAAAUUGCAUUUCCAUUUCAGCGC__AAAGA_______AAAGAAAACGAAAAGUGGAAUGCAAUUUGCUCGAAUGCAUUUAACUCAAAAAU__UUCAUGAAAAUGCAACAAAAUC .(((.((((((((((((((.((((............................)))).).)))))))))))))......)))....................................... (-14.36 = -14.89 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:38 2006