
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,734,487 – 5,734,597 |
| Length | 110 |
| Max. P | 0.988789 |

| Location | 5,734,487 – 5,734,597 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -28.59 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.53 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5734487 110 + 22224390 UGCGCAUUCCCAUUUGGGGUGGCAAUUGUAACGGCACCCACCCCUAUAUCGUU---ACC---UACACCCUGUAUAUCCUUCACAUCCGGGUGAAUCAUUCAGCGCCUGCUUUUAUC .((((..........(((((((....(((....))).))))))).........---...---..((((((((.........)))...))))).........))))........... ( -28.60) >DroSim_CAF1 8213 109 + 1 UGCGCAUUCCCAUUUGGGGUGGCAAUUGUAACGGCACCCACCCCUAUAUCGU----ACC---UACACCCUGUAUAUCCUUCACAUCCGGGUGAAUCAUUCAGCGCCUGCUUUUAUC .((((..........(((((((....(((....))).)))))))........----...---..((((((((.........)))...))))).........))))........... ( -28.60) >DroEre_CAF1 7974 105 + 1 UGCGCAUUCCCAGUUGGGGUGGCAAUUGUAAGGGCACCCACCCCUAUAUCGU-----------AUACCCUGUAUAUCCUUCACAUCCGGGUGAAUCAUUCAGCGCCUGCUUUAAUU .((((...(((....(((((((....(((....))).)))))))......((-----------((((...))))))...........)))(((.....)))))))........... ( -27.50) >DroYak_CAF1 8889 114 + 1 UGCGCAUUCCCAGUUGGGGUGGCAAUUGUAACGGCACCCACCCCUAUGCCAUGC--ACCCUAUAUAUCCUGCAUAUCCUUCACAUCCGGGUGAAUCAUUCAGCGCCUGCUUUUAUU .((((...(((.((.(((((((....(((....))).)))))))...)).((((--(............))))).............)))(((.....)))))))........... ( -29.00) >DroAna_CAF1 6976 93 + 1 UGAGCAUUCCCAUUUGGGGUGGUAAUUG-------AGCCACCCCUAUGUCUUAUAAACU---CACACCC-AAAUAUCCUCGAUAUCCGGGUGAAUCAUUUUACU------------ ((((......(((..((((((((.....-------.)))))))).))).........))---))(((((-..(((((...)))))..)))))............------------ ( -29.26) >consensus UGCGCAUUCCCAUUUGGGGUGGCAAUUGUAACGGCACCCACCCCUAUAUCGU____ACC___UACACCCUGUAUAUCCUUCACAUCCGGGUGAAUCAUUCAGCGCCUGCUUUUAUC .((((..........(((((((....(((....))).)))))))....................(((((..................))))).........))))........... (-23.60 = -23.53 + -0.07)

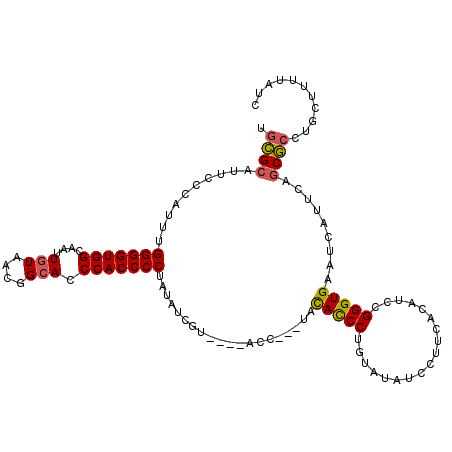

| Location | 5,734,487 – 5,734,597 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5734487 110 - 22224390 GAUAAAAGCAGGCGCUGAAUGAUUCACCCGGAUGUGAAGGAUAUACAGGGUGUA---GGU---AACGAUAUAGGGGUGGGUGCCGUUACAAUUGCCACCCCAAAUGGGAAUGCGCA ...........((((.........(((((..((((.....))))...)))))..---...---.........(((((((.((......))....)))))))..........)))). ( -31.20) >DroSim_CAF1 8213 109 - 1 GAUAAAAGCAGGCGCUGAAUGAUUCACCCGGAUGUGAAGGAUAUACAGGGUGUA---GGU----ACGAUAUAGGGGUGGGUGCCGUUACAAUUGCCACCCCAAAUGGGAAUGCGCA ...........((((.........(((((..((((.....))))...)))))..---...----........(((((((.((......))....)))))))..........)))). ( -31.20) >DroEre_CAF1 7974 105 - 1 AAUUAAAGCAGGCGCUGAAUGAUUCACCCGGAUGUGAAGGAUAUACAGGGUAU-----------ACGAUAUAGGGGUGGGUGCCCUUACAAUUGCCACCCCAACUGGGAAUGCGCA ...........(((((((.....)))(((((.((((.......))))((((..-----------.((((.(((((((....)))))))..))))..))))...)))))...)))). ( -30.50) >DroYak_CAF1 8889 114 - 1 AAUAAAAGCAGGCGCUGAAUGAUUCACCCGGAUGUGAAGGAUAUGCAGGAUAUAUAGGGU--GCAUGGCAUAGGGGUGGGUGCCGUUACAAUUGCCACCCCAACUGGGAAUGCGCA ...........(((((((.....)))(((((.((((.....((((((............)--))))).))))(((((((.((......))....)))))))..)))))...)))). ( -35.60) >DroAna_CAF1 6976 93 - 1 ------------AGUAAAAUGAUUCACCCGGAUAUCGAGGAUAUUU-GGGUGUG---AGUUUAUAAGACAUAGGGGUGGCU-------CAAUUACCACCCCAAAUGGGAAUGCUCA ------------............((((((((((((...)))))))-)))))((---(((........(((.(((((((..-------......)))))))..))).....))))) ( -33.62) >consensus AAUAAAAGCAGGCGCUGAAUGAUUCACCCGGAUGUGAAGGAUAUACAGGGUGUA___GGU____ACGAUAUAGGGGUGGGUGCCGUUACAAUUGCCACCCCAAAUGGGAAUGCGCA ...........((((.........(((((..((((.....))))...)))))....................(((((((...............)))))))..........)))). (-26.04 = -26.20 + 0.16)

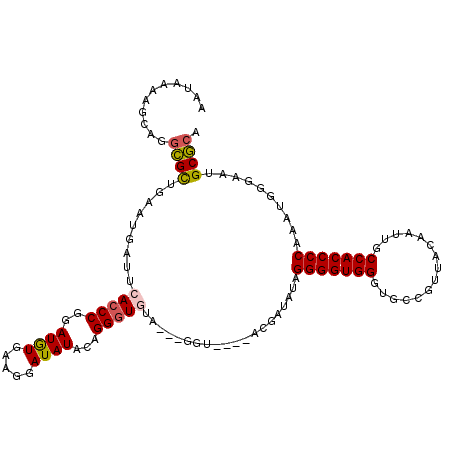
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:36 2006