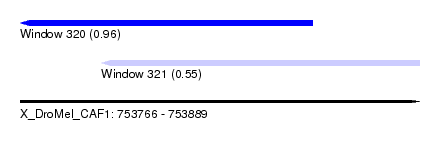
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 753,766 – 753,889 |
| Length | 123 |
| Max. P | 0.960235 |

| Location | 753,766 – 753,856 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.27 |
| Mean single sequence MFE | -18.75 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 753766 90 - 22224390 GCAA-ACCC------AAACAC-------CCAACCCCCAAGAAAAUCCUUUCGCCCGGGGA-ACUACAGUACACUGCUGCACUUAGCCAAGUUAAGUUAACCCAAA (((.-....------......-------....((((...((((....))))....)))).-....(((....))).)))(((((((...)))))))......... ( -17.20) >DroSec_CAF1 68781 91 - 1 GCAA-ACCC------AAACAC-------CCAACCCCCAAGAAAAUCCUUUCGCCCGGGGGCGAAACAGUGCACUGCUGCACUUAGCCAAGUUUAGUUAACCCAAA ...(-((..------((((..-------....(((....((((....))))....)))(((.....((((((....))))))..)))..)))).)))........ ( -20.30) >DroEre_CAF1 75690 104 - 1 CCAAACUCCAAACUCCAACCCCCAUGUCGCAAGCCCCAAGAAAAUCCUUUCGCCCGGGGA-ACUACAGUGCACUGCUGCACUUAGCCAAGUUAAGUUAACCCAAA ...((((...((((.....((((....(....)......((((....))))....)))).-.(((.((((((....)))))))))...)))).))))........ ( -19.80) >DroYak_CAF1 71765 90 - 1 GCAA-CUCC------CAACCC-------UCAAACCCCAAGAAAAUCCUUUCGCCCGGGGA-ACUACAGUGCACUGCUGCACUUAGCCAAGUUAAGUUAACCCAAA ..((-((..------.(((..-------.....((((..((((....))))....)))).-.(((.((((((....)))))))))....))).))))........ ( -17.70) >consensus GCAA_ACCC______AAACAC_______CCAACCCCCAAGAAAAUCCUUUCGCCCGGGGA_ACUACAGUGCACUGCUGCACUUAGCCAAGUUAAGUUAACCCAAA (((..............................((((..((((....))))....))))......(((....))).)))(((((((...)))))))......... (-14.32 = -14.82 + 0.50)

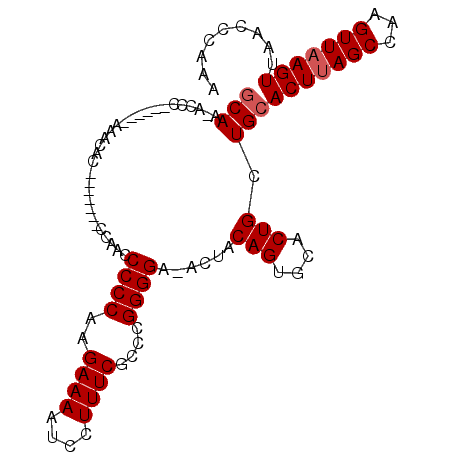
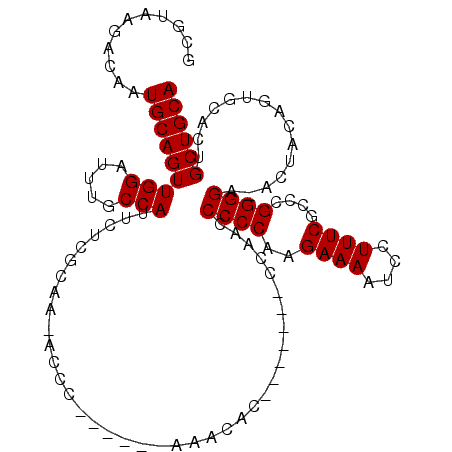
| Location | 753,791 – 753,889 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.99 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -17.17 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 753791 98 - 22224390 GCGUAAGACAAUGCAGUUGGAUUUGCCAUUCUCGCAA-ACCC------AAACAC-------CCAACCCCCAAGAAAAUCCUUUCGCCCGGGGA-ACUACAGUACACUGCUGCA ...........(((((((((.(((((.......))))-).))------))....-------....((((...((((....))))....)))).-....(((....)))))))) ( -23.70) >DroSec_CAF1 68806 99 - 1 GCGUAAGACAAUGCAGUUGGAUUUGCCAUUCUUGCAA-ACCC------AAACAC-------CCAACCCCCAAGAAAAUCCUUUCGCCCGGGGGCGAAACAGUGCACUGCUGCA ..(((.(.((.((((.((((.(((((.......))))-).))------))....-------....(((((..((((....))))....)))))........)))).)))))). ( -26.50) >DroEre_CAF1 75715 112 - 1 GCGUAAGACAAUGCAGUUGGAUUUGCCAUUCUCCCAAACUCCAAACUCCAACCCCCAUGUCGCAAGCCCCAAGAAAAUCCUUUCGCCCGGGGA-ACUACAGUGCACUGCUGCA (((((.((((.((..((((((((((................)))).))))))...))))))(((..((((..((((....))))....)))).-.......)))..))).)). ( -25.09) >DroYak_CAF1 71790 98 - 1 GCGUAAGACAAUGCAGUUGGAUUUGCCAUACUCGCAA-CUCC------CAACCC-------UCAAACCCCAAGAAAAUCCUUUCGCCCGGGGA-ACUACAGUGCACUGCUGCA ..(((.(.((.(((((((((..((((.......))))-...)------))))..-------.....((((..((((....))))....)))).-.......)))).)))))). ( -23.90) >consensus GCGUAAGACAAUGCAGUUGGAUUUGCCAUUCUCGCAA_ACCC______AAACAC_______CCAACCCCCAAGAAAAUCCUUUCGCCCGGGGA_ACUACAGUGCACUGCUGCA ...........(((((((((.....)))......................................((((..((((....))))....))))...............)))))) (-17.17 = -17.18 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:06 2006