
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,707,435 – 5,707,591 |
| Length | 156 |
| Max. P | 0.996724 |

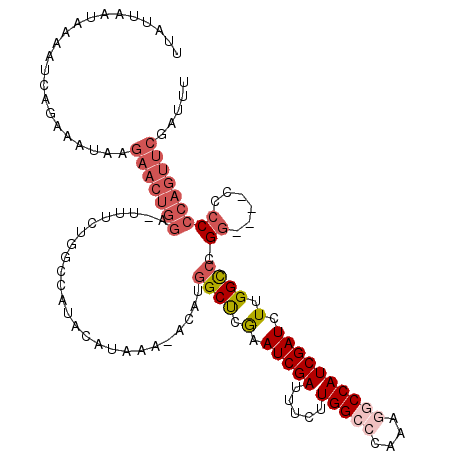
| Location | 5,707,435 – 5,707,551 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -16.78 |
| Energy contribution | -18.82 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5707435 116 + 22224390 ---UUAAUGAAAUCAGAAAUAAGAACUAGA-UUUCUUGCCAUACACAAAUACAUGGCUCGAAUCGAUAUCUUGGCCCAAAGGCCAUCGAUCUUGGUCCGGGCCCCCCCCCAGUUCGAUUU ---......((((((....)..(((((.((-(((...(((((..........)))))..)))))........(((((...(((((.......))))).))))).......)))))))))) ( -31.00) >DroSec_CAF1 35642 115 + 1 UUGUUAAUAUAAUCAGAAAUAAGAACUGGA-UUUCUGGCCAUACAUAAAUACAUGGCUCGAAUCGAUUCCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG----CCCCCCAGUUCGAUUU ......................(((((((.-.(((.((((((..........)))))).)))..........((((....(((((.......))))).))----))..)))))))..... ( -34.80) >DroSim_CAF1 33240 97 + 1 UUGUUAAUAUAAUCAGAAAUAAGAACUGGA-UUUCUG------------------GCUCGAAUCGAUUCCUUGGCCCACAGGCCAUCGAUCUUGGCCCGG----CCCCCCAGUUCGAUUU ......................(((((((.-...(((------------------(..((((((((.....(((((....)))))))))).)))..))))----....)))))))..... ( -30.50) >DroEre_CAF1 26411 113 + 1 UUAUUAAUAAAACCAGAACCAAUUUCUGGACUUUCUGGCCAUACAUAAA-AGAUGGCC-AAAUCGAUUUCAUGGCCCAAACACCAUCGAUCUUGGCCUGC-----CACCCAGUUCGAUUU ............((((((.....))))))......(((((((.(.....-.)))))))-)((((((.....((((((((............))))...))-----))......)))))). ( -28.40) >DroYak_CAF1 27785 113 + 1 UUACUAAUAAAUUCAGAACUAGGAACUGCACUUUCUGGCCGUACAUAA--ACAUGGCCGGAAUCGAUUUCGUGGCCCAAGAACCAUCGAUCUUGGCCUGG-----CCCCCCAUUCGAUUU ................................((((((((((......--..))))))))))((((......((((.((((........))))))))(((-----....))).))))... ( -28.20) >consensus UUAUUAAUAAAAUCAGAAAUAAGAACUGGA_UUUCUGGCCAUACAUAAA_ACAUGGCUCGAAUCGAUUUCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG____CCCCCCAGUUCGAUUU ......................(((((((.........................((((.(.(((((.....(((((....)))))))))).).)))).((......)))))))))..... (-16.78 = -18.82 + 2.04)


| Location | 5,707,435 – 5,707,551 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.94 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5707435 116 - 22224390 AAAUCGAACUGGGGGGGGGCCCGGACCAAGAUCGAUGGCCUUUGGGCCAAGAUAUCGAUUCGAGCCAUGUAUUUGUGUAUGGCAAGAAA-UCUAGUUCUUAUUUCUGAUUUCAUUAA--- .((((((.(((((......)))))......(((..(((((....))))).))).))))))...((((((........))))))..((((-((.((.........)))))))).....--- ( -36.10) >DroSec_CAF1 35642 115 - 1 AAAUCGAACUGGGGGG----CCGGGCCAAGAUCGAUGGCCUUUGGGCCAAGGAAUCGAUUCGAGCCAUGUAUUUAUGUAUGGCCAGAAA-UCCAGUUCUUAUUUCUGAUUAUAUUAACAA .....((((((((.((----((.(((...(((((((((((....))))).....))))))...)))..((((....)))))))).....-))))))))...................... ( -38.90) >DroSim_CAF1 33240 97 - 1 AAAUCGAACUGGGGGG----CCGGGCCAAGAUCGAUGGCCUGUGGGCCAAGGAAUCGAUUCGAGC------------------CAGAAA-UCCAGUUCUUAUUUCUGAUUAUAUUAACAA .....((((((((...----...(((...(((((((((((....))))).....))))))...))------------------).....-))))))))...................... ( -31.30) >DroEre_CAF1 26411 113 - 1 AAAUCGAACUGGGUG-----GCAGGCCAAGAUCGAUGGUGUUUGGGCCAUGAAAUCGAUUU-GGCCAUCU-UUUAUGUAUGGCCAGAAAGUCCAGAAAUUGGUUCUGGUUUUAUUAAUAA .(((.((((..(.((-----(....))).(((((((....(((((((..((....)).(((-(((((((.-.....).)))))))))..))))))).))))))))..)))).)))..... ( -33.50) >DroYak_CAF1 27785 113 - 1 AAAUCGAAUGGGGGG-----CCAGGCCAAGAUCGAUGGUUCUUGGGCCACGAAAUCGAUUCCGGCCAUGU--UUAUGUACGGCCAGAAAGUGCAGUUCCUAGUUCUGAAUUUAUUAGUAA ...((((((.(((((-----(..((((..(((((((((((....))))).....))))))..))))....--...(((((.........))))))))))).)))).))............ ( -34.20) >consensus AAAUCGAACUGGGGGG____CCGGGCCAAGAUCGAUGGCCUUUGGGCCAAGAAAUCGAUUCGAGCCAUGU_UUUAUGUAUGGCCAGAAA_UCCAGUUCUUAUUUCUGAUUUUAUUAACAA .....(((((((((......)).(((...(((((((((((....))))).....))))))...))).........................)))))))...................... (-21.34 = -21.94 + 0.60)


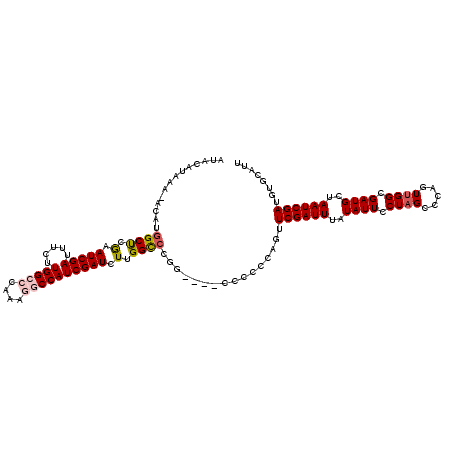
| Location | 5,707,471 – 5,707,591 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.95 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5707471 120 + 22224390 AUACACAAAUACAUGGCUCGAAUCGAUAUCUUGGCCCAAAGGCCAUCGAUCUUGGUCCGGGCCCCCCCCCAGUUCGAUUUAUAUUCCUAGCCCAGUUGGCGAUGCUAAUCGAUGUGCAUU ...((((.......((((((.(((((.(((.(((((....)))))..))).))))).))))))..........((((((..((((.(((((...))))).))))..)))))))))).... ( -38.70) >DroSec_CAF1 35681 116 + 1 AUACAUAAAUACAUGGCUCGAAUCGAUUCCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG----CCCCCCAGUUCGAUUUAUAUUCCUAGUCCAGUUGGCGAUGCUAAUCGAUGUGCAUU .(((((..((...(((((((((((((......((((....(((((.......))))).))----)).......))))))).........(((.....))))).))))))..))))).... ( -29.72) >DroSim_CAF1 33276 101 + 1 ---------------GCUCGAAUCGAUUCCUUGGCCCACAGGCCAUCGAUCUUGGCCCGG----CCCCCCAGUUCGAUUUAUAUUCCUAGCCCAGUUGGCGAUGCUAAUCGAUGUGCAUU ---------------((..(((((((......((((....(((((.......))))).))----)).......)))))))(((((....(((.....)))(((....))))))))))... ( -29.02) >DroEre_CAF1 26451 113 + 1 AUACAUAAA-AGAUGGCC-AAAUCGAUUUCAUGGCCCAAACACCAUCGAUCUUGGCCUGC-----CACCCAGUUCGAUUUAUAUUCCUCGCCCAGUUGGCGAUGCUAAUCGAUGUGCAUU .........-....((((-(((((((.....(((........)))))))).))))))(((-----.((.....((((((..((....(((((.....)))))))..)))))).))))).. ( -28.30) >DroYak_CAF1 27825 113 + 1 GUACAUAA--ACAUGGCCGGAAUCGAUUUCGUGGCCCAAGAACCAUCGAUCUUGGCCUGG-----CCCCCCAUUCGAUUUAUAUUCCUAGCCCAUUUGGCGAUGCUAAUCGAUGUGCAUU (((((...--....((((((..((((..((((((........))).)))..)))).))))-----))......((((((..((((....(((.....)))))))..)))))))))))... ( -32.40) >consensus AUACAUAAA_ACAUGGCUCGAAUCGAUUUCUUGGCCCAAAGGCCAUCGAUCUUGGCCCGG____CCCCCCAGUUCGAUUUAUAUUCCUAGCCCAGUUGGCGAUGCUAAUCGAUGUGCAUU ..............((((.(.(((((.....(((((....)))))))))).).))))................((((((..((((.((((.....)))).))))..))))))........ (-21.60 = -22.24 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:16 2006