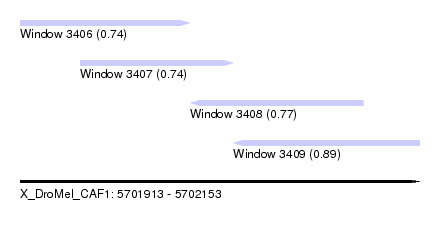
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,701,913 – 5,702,153 |
| Length | 240 |
| Max. P | 0.892426 |

| Location | 5,701,913 – 5,702,015 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -20.73 |
| Energy contribution | -20.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5701913 102 + 22224390 AUUCAAAUUAGUCCAAGCGAGCCCCACUAAC-CGAUGCGGAAAAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCC ..................(((...(((.((.-((((((((.....))..((((((((((((((......)))))))..))))))))))))).)))))..))). ( -22.00) >DroSec_CAF1 23148 101 + 1 AUUCAAAUUAGGCCAAGCGAGCUCCACUAAC-CGAUGCGGA-AAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCC .........(((((((.(((((((.......-......((.-...))....((((((((((((......)))))))..))))))))).))).))).))))... ( -23.40) >DroSim_CAF1 26452 101 + 1 AUUCAAAUUAGGCCAAGCGAGCUCCACUAAC-CGAUGCGGA-AAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCC .........(((((((.(((((((.......-......((.-...))....((((((((((((......)))))))..))))))))).))).))).))))... ( -23.40) >DroEre_CAF1 21102 99 + 1 AUUCAAAUUUGGCCAAUC---CCCCACCCACUCGAUGCGGU-AAUCCAAUUGAAUUUCUAUUGAUUCCUCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCC .........(((......---..)))..((((((((((((.-...))..(((((((((((((((....))))))))..))))))))))))))..)))...... ( -23.70) >consensus AUUCAAAUUAGGCCAAGCGAGCCCCACUAAC_CGAUGCGGA_AAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCC ........................(((.....((((((((.....))..((((((((((((((......)))))))..)))))))))))))...)))...... (-20.73 = -20.72 + -0.00)

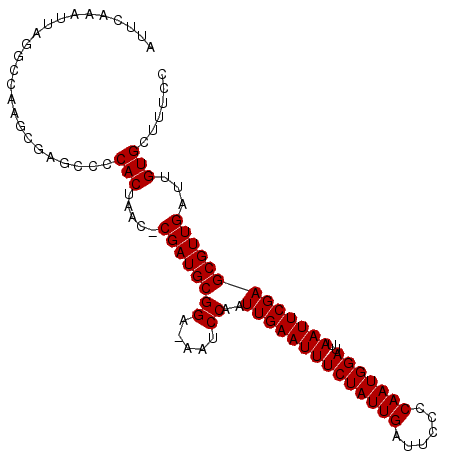

| Location | 5,701,949 – 5,702,041 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 97.09 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5701949 92 + 22224390 CGGAAAAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCCAUAUAUUUGCGCUUUAGUUGCGGUAC ......((((((((((((((((((((......)))))))..)))))((((((.(((((((.....)))).))).)))))).))))).))).. ( -21.60) >DroSec_CAF1 23184 91 + 1 CGGA-AAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCCAUAUAUUUGCGCUUUAGUUGCGCUAC .(((-((.(((((..(..(((.((((((((......))))))))..)))..)..)))).).)))))........((((.......))))... ( -23.30) >DroSim_CAF1 26488 91 + 1 CGGA-AAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCCAUAUAUUUGCGCUUUAGUUGCGCUAC .(((-((.(((((..(..(((.((((((((......))))))))..)))..)..)))).).)))))........((((.......))))... ( -23.30) >DroEre_CAF1 21136 91 + 1 CGGU-AAUCCAAUUGAAUUUCUAUUGAUUCCUCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCCAUAUAUUUGCGCUUUAGCUGCGGUAC ....-...((..(((((((((((((((....))))))))..)))))))(((((((..((((.............)))).))))).))))... ( -22.72) >consensus CGGA_AAUCCAAUUGAAUUUCUAUUGAUUCCCCAAUGGAUUAAUUCGAGCGUUGAUUGUGCUUUCCAUAUAUUUGCGCUUUAGUUGCGCUAC .((.....))....((((((((((((......)))))))..)))))((((((.(((((((.....)))).))).))))))............ (-19.70 = -19.70 + -0.00)


| Location | 5,702,015 – 5,702,119 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -20.05 |
| Energy contribution | -22.55 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5702015 104 - 22224390 CAGACAGCGAUAAAUCAAUAUCAUGCGAGUAUGUAUGCCGGUUGUGGGUACAUAAUUCACACAUAGCCAGGCCGCACUGUACCGCAACUAAAGCGCAAAUAUAU ......((((((......)))).((((.((((((..((((((((((.((.........)).))))))).)))...)).))))))))........))........ ( -26.00) >DroSec_CAF1 23249 103 - 1 CAGACAGCGAUAAAUCAAUAUCAUGCGCGUAUGUCUGCCGGCUGUGUGUACAUA-UUCGCACAUAGCCAGGCCGCACUGUAGCGCAACUAAAGCGCAAAUAUAU ......((((((......)))).(((((....(((.(((((((((((((.....-...)))))))))).))).).))....)))))........))........ ( -36.30) >DroSim_CAF1 26553 103 - 1 CAGACAGCGAUAAAUCAAUAUCAUGCGAGUAUGUCUGCCGGCUGUGUGUACAUA-UUCGCACAUAGCCAGGCCGCACUGUAGCGCAACUAAAGCGCAAAUAUAU ...((((.((((......)))).((((((.....))(((((((((((((.....-...)))))))))).))))))))))).((((.......))))........ ( -36.20) >DroEre_CAF1 21201 97 - 1 CAGACGGCGAUAAAUCAAUAACCUGCGAGCAUGUAU-------GUAUGUACGCCAUGUGCACAUAGCCAGGCCGCACUGUACCGCAGCUAAAGCGCAAAUAUAU ......(((..............((((.((.((..(-------((.(((((.....))))).)))..)).))))))(((.....)))......)))........ ( -21.10) >consensus CAGACAGCGAUAAAUCAAUAUCAUGCGAGUAUGUAUGCCGGCUGUGUGUACAUA_UUCGCACAUAGCCAGGCCGCACUGUACCGCAACUAAAGCGCAAAUAUAU ...((((.((((......)))).((((.........(((((((((((((.........)))))))))).))))))))))).((((.......))))........ (-20.05 = -22.55 + 2.50)

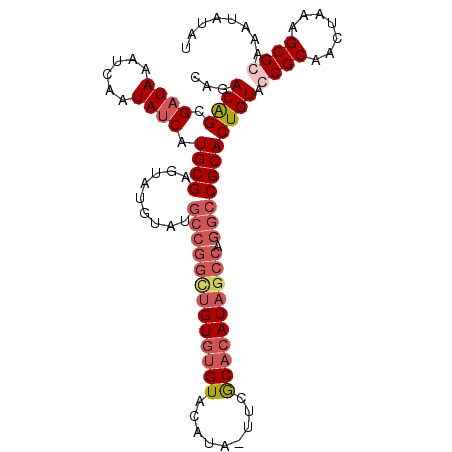
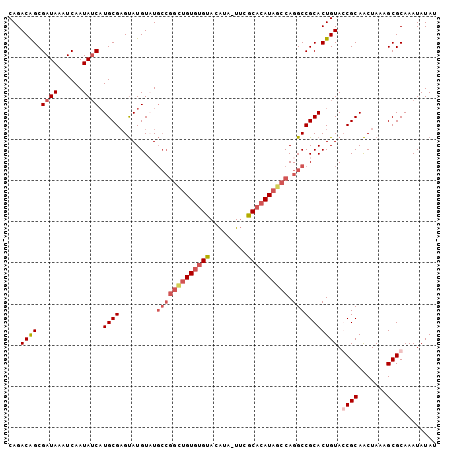
| Location | 5,702,041 – 5,702,153 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.12 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -27.46 |
| Energy contribution | -28.78 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5702041 112 - 22224390 GGCUAAUUGUUUCUGGCCAAAUAAAUCUGUGCACCAGACAGCGAUAAAUCAAUAUCAUGCGAGUAUGUAUGCCGGUUGUGGGUACAUAAUUCACACAUAGCCAGGCCGCACU (((((........)))))..........((((..((.((.((((((......))))..))..)).))...((((((((((.((.........)).))))))).))).)))). ( -28.20) >DroSec_CAF1 23275 111 - 1 GGCUAAUUGUUUCUGGCCAAAUAAAUCUGUGCGCCAGACAGCGAUAAAUCAAUAUCAUGCGCGUAUGUCUGCCGGCUGUGUGUACAUA-UUCGCACAUAGCCAGGCCGCACU (((((........)))))..........(((((((((((((((................)))...)))))...((((((((((.....-...)))))))))).)).))))). ( -40.39) >DroSim_CAF1 26579 111 - 1 GGCUAAUUGUUUCUGGCCAAAUAAAUCUGUGCGCCAGACAGCGAUAAAUCAAUAUCAUGCGAGUAUGUCUGCCGGCUGUGUGUACAUA-UUCGCACAUAGCCAGGCCGCACU (((((........)))))..........((((((((((((((((((......))))..)).....)))))...((((((((((.....-...)))))))))).)).))))). ( -39.80) >DroEre_CAF1 21227 105 - 1 GGCUAAUUGUUUCUGGCCAAAUAAAUCUGUGCGCCAGACGGCGAUAAAUCAAUAACCUGCGAGCAUGUAU-------GUAUGUACGCCAUGUGCACAUAGCCAGGCCGCACU ((((.......((((((((..........)).)))))).(((................(((.(((((...-------.))))).))).(((....))).))).))))..... ( -28.30) >consensus GGCUAAUUGUUUCUGGCCAAAUAAAUCUGUGCGCCAGACAGCGAUAAAUCAAUAUCAUGCGAGUAUGUAUGCCGGCUGUGUGUACAUA_UUCGCACAUAGCCAGGCCGCACU ((((.((((((((((((...(((....)))..)))))).))))))..........((((....))))......((((((((((.........)))))))))).))))..... (-27.46 = -28.78 + 1.31)

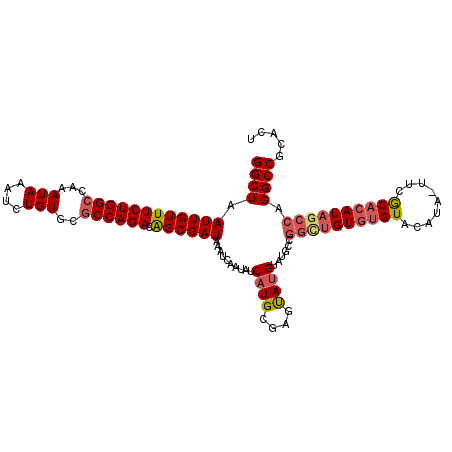

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:09 2006