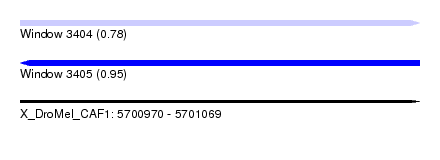
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,700,970 – 5,701,069 |
| Length | 99 |
| Max. P | 0.951244 |

| Location | 5,700,970 – 5,701,069 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -13.10 |
| Energy contribution | -12.91 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
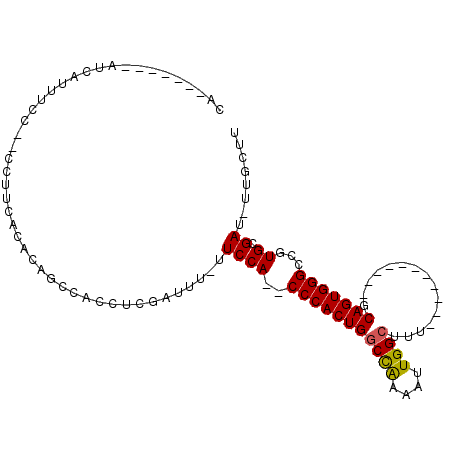
>X_DroMel_CAF1 5700970 99 + 22224390 CA-------AUCAUUUCCCCCCUUCACACAGCCAUCUCGAUUUUUUCCA--CCCACUGGCUAAAAUUGGCUUUAGCCAACUGCUGCAGUGGGCCGUGCGAUUUUGCUU ..-------..............((.(((.(((.....((.....))..--..(((((.(.....(((((....))))).....))))))))).))).))........ ( -22.80) >DroSec_CAF1 22186 84 + 1 CA-------AUCAUUUCC--CCUUCACACAGCCACCUCGAUUU-UUCCA--CCCACUGGCCAAAAUUGGCUUU-----------GCAGUGGGCCGUGCGAU-UUGCUU ..-------.........--...((.(((.(((.....((...-.))..--..(((((((((....))))...-----------.)))))))).))).)).-...... ( -19.80) >DroSim_CAF1 25498 84 + 1 CA-------AUCAUUUCC--CCUUCACACAGCCACCUCGAUUU-UUCCA--CCCACUGGCCAAAAUUGGCUUU-----------GCAGUGGGCCGUGCGAU-UUGCUU ..-------.........--...((.(((.(((.....((...-.))..--..(((((((((....))))...-----------.)))))))).))).)).-...... ( -19.80) >DroEre_CAF1 20141 96 + 1 CAAUCUGCAAUCAUAU-------CCAACCAACCACCUCGAUUU-UUCCACACCCACUGACUGAAAUUGGCUUUAGCCGCC---UGCAGUGGGGCGUGCGAU-UUGCUU ......((((((....-------...............((...-.))(((.(((((((.(......((((....))))..---.))))))))..))).)).-)))).. ( -23.10) >consensus CA_______AUCAUUUCC__CCUUCACACAGCCACCUCGAUUU_UUCCA__CCCACUGGCCAAAAUUGGCUUU___________GCAGUGGGCCGUGCGAU_UUGCUU .............................................((((..(((((((((((....))))...............)))))))...)).))........ (-13.10 = -12.91 + -0.19)


| Location | 5,700,970 – 5,701,069 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.86 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
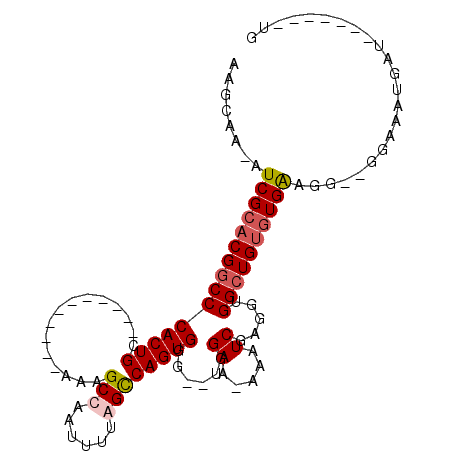
>X_DroMel_CAF1 5700970 99 - 22224390 AAGCAAAAUCGCACGGCCCACUGCAGCAGUUGGCUAAAGCCAAUUUUAGCCAGUGGG--UGGAAAAAAUCGAGAUGGCUGUGUGAAGGGGGGAAAUGAU-------UG ........((((((((((((((((...(((((((....)))))))...).)))))((--(.......))).....))))))))))..............-------.. ( -32.50) >DroSec_CAF1 22186 84 - 1 AAGCAA-AUCGCACGGCCCACUGC-----------AAAGCCAAUUUUGGCCAGUGGG--UGGAA-AAAUCGAGGUGGCUGUGUGAAGG--GGAAAUGAU-------UG ......-.(((((((((((((((.-----------...((((....)))))))))((--(....-..))).....))))))))))...--.........-------.. ( -28.20) >DroSim_CAF1 25498 84 - 1 AAGCAA-AUCGCACGGCCCACUGC-----------AAAGCCAAUUUUGGCCAGUGGG--UGGAA-AAAUCGAGGUGGCUGUGUGAAGG--GGAAAUGAU-------UG ......-.(((((((((((((((.-----------...((((....)))))))))((--(....-..))).....))))))))))...--.........-------.. ( -28.20) >DroEre_CAF1 20141 96 - 1 AAGCAA-AUCGCACGCCCCACUGCA---GGCGGCUAAAGCCAAUUUCAGUCAGUGGGUGUGGAA-AAAUCGAGGUGGUUGGUUGG-------AUAUGAUUGCAGAUUG ......-.((.((((((((((((.(---(..(((....)))..)).))))..).))))))))).-.((((...(..((((.....-------...))))..).)))). ( -27.10) >consensus AAGCAA_AUCGCACGGCCCACUGC___________AAAGCCAAUUUUAGCCAGUGGG__UGGAA_AAAUCGAGGUGGCUGUGUGAAGG__GGAAAUGAU_______UG ........(((((((((((((((...............(((......))))))))......((.....)).....))))))))))....................... (-16.99 = -17.86 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:05 2006