
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,696,945 – 5,697,193 |
| Length | 248 |
| Max. P | 0.990250 |

| Location | 5,696,945 – 5,697,062 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -14.12 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5696945 117 - 22224390 GCGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUUUCUGCCUUUGCGCUCUUGUUUUUC-UCGCACCCGCGCCCGCGCCAUUCC ((((...((..(((...((((((....))))))..)))..))((((((.....)))))--)...................((((...(((.....-..)))...))))..))))...... ( -23.20) >DroSec_CAF1 17894 111 - 1 GCGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUUUUUUUUCUGCCUUUGCGCUCUUGUUUUUC-CCGCACCC------CCGCCAUUCC ((((...((..(((...((((((....))))))..)))..))((((((.....)))))--)...................))))...........-..((....------..))...... ( -18.20) >DroSim_CAF1 21168 111 - 1 GCGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA--UUUUUAUUUUUUCUGCCUUUGCGCUCUUGUUCUUC-CCGCACCC------CCGCCAUUCC ((((...((..(((...((((((....))))))..)))..))((((((.....)))))--)...................))))...........-..((....------..))...... ( -18.20) >DroEre_CAF1 17104 113 - 1 -CGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAAAUUUUUUGUUUUUUCCGCCCUUGCGCUCUUGUUUUUCGCCGACC------CGUCGCCAUUCC -(((...((..(((...((((((....))))))..)))..)).(((((.....)))))......................)))............(.(((..------..))).)..... ( -15.30) >DroYak_CAF1 20805 115 - 1 -CGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA---UUUUAUUUUUUCUGCCUUGGCGCUCUUGUUUUUU-CCGCUCCCUCGCCAUUGCCAUUCC -......((..(((...((((((....))))))..)))..))((((((.....)))))---)............((..(((((.....((.....-..)).....)))))..))...... ( -19.90) >consensus GCGCUUCGCUUGUGUUUAUGCAAAUAUUUGUAUUCCAUUUGCAUUUCAAUAAUUGAAA__UUUUUAUUUUUUCUGCCUUUGCGCUCUUGUUUUUC_CCGCACCC____C_CCGCCAUUCC ((((...((..(((...((((((....))))))..)))..)).(((((.....)))))......................)))).................................... (-14.12 = -14.52 + 0.40)

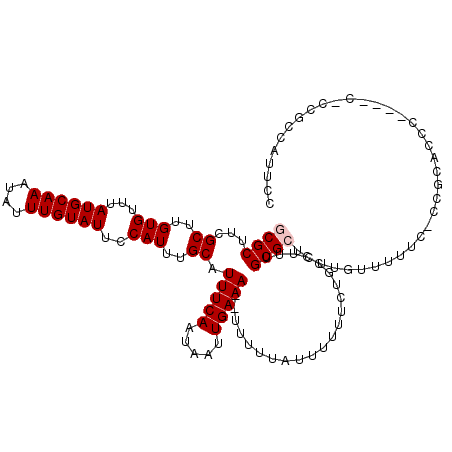

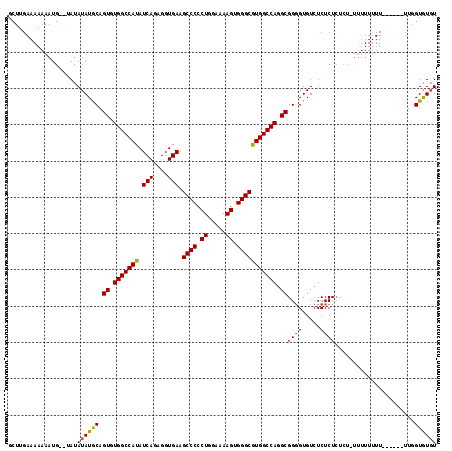
| Location | 5,697,062 – 5,697,180 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -27.63 |
| Energy contribution | -27.63 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5697062 118 - 22224390 GCUCGAAAAAAAUGAAUAUAUAUACUGUGUGGCCAUAUCAGAGGUGAAGCCCUCUAGAAAAGUGGGCGUGGCCAGGCGGUGUGUCUCUCUCUCUCUUUUUUUUUUUUUGUUCGUAUUU (..(((((((((.(((...(((((((((.(((((((.(((....))).((((.((.....)).))))))))))).)))))))))............))).)))))))))..)...... ( -37.16) >DroSec_CAF1 18005 102 - 1 GCUUGAAAAAAAUG--UAUAUAUGCAGUGUGGCCAUAUCAGAGGUGAAGCCCCCUGGAAAAGUGGGCAUGGCCAGGCGGGGUGUCUCUCU--------UUUUU------UUGGUGUGU (((.(((((((..(--...((((.(.((.(((((((.(((....))).((((.((.....)).))))))))))).)).).))))..)..)--------)))))------).))).... ( -33.10) >DroSim_CAF1 21279 110 - 1 GCUUGAAAAAAAUG--UAUAUAUGCAGUGUGGCCAUAUCAGAGGUGAAGCCCCCUGGAAAAGUGGGCGUGGCCAGGCGGGGUGUCUCUCUCUCUUUUUUUUUU------UUGGUGUGU (((.((((((((((--((....))))((.(((((((.(((....))).((((.((.....)).))))))))))).))((((.(.....).))))..)))))))------).))).... ( -33.10) >consensus GCUUGAAAAAAAUG__UAUAUAUGCAGUGUGGCCAUAUCAGAGGUGAAGCCCCCUGGAAAAGUGGGCGUGGCCAGGCGGGGUGUCUCUCUCUCU_UUUUUUUU______UUGGUGUGU ...................((((((.((.(((((((.(((....))).((((.((.....)).))))))))))).))((((.....))))......................)))))) (-27.63 = -27.63 + 0.01)


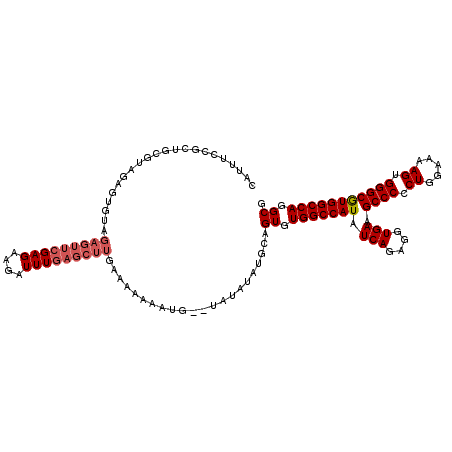
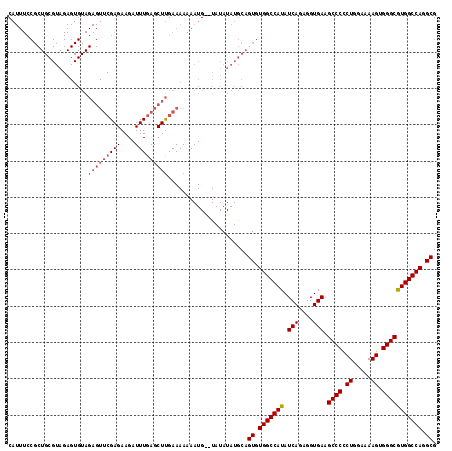
| Location | 5,697,102 – 5,697,193 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -26.45 |
| Energy contribution | -28.23 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5697102 91 - 22224390 C---------------------------GAGAAGAUUUGAGCUCGAAAAAAAUGAAUAUAUAUACUGUGUGGCCAUAUCAGAGGUGAAGCCCUCUAGAAAAGUGGGCGUGGCCAGGCG (---------------------------(((..........)))).....................((.(((((((.(((....))).((((.((.....)).))))))))))).)). ( -27.80) >DroSec_CAF1 18031 116 - 1 CAUUUCCGCUGCGUAGAGUGUAGAGUUCGAGAAGAUUUGAGCUUGAAAAAAAUG--UAUAUAUGCAGUGUGGCCAUAUCAGAGGUGAAGCCCCCUGGAAAAGUGGGCAUGGCCAGGCG ......(((((((((..((...(((((((((....))))))))).......)).--....)))))))))(((((((.(((....))).((((.((.....)).))))))))))).... ( -38.40) >DroSim_CAF1 21313 116 - 1 CAUUUCCGCUGCGUAGAGUGUAGAGUUCGAGAAGAUUUGAGCUUGAAAAAAAUG--UAUAUAUGCAGUGUGGCCAUAUCAGAGGUGAAGCCCCCUGGAAAAGUGGGCGUGGCCAGGCG ......(((((((((..((...(((((((((....))))))))).......)).--....)))))))))(((((((.(((....))).((((.((.....)).))))))))))).... ( -38.70) >consensus CAUUUCCGCUGCGUAGAGUGUAGAGUUCGAGAAGAUUUGAGCUUGAAAAAAAUG__UAUAUAUGCAGUGUGGCCAUAUCAGAGGUGAAGCCCCCUGGAAAAGUGGGCGUGGCCAGGCG ......................(((((((((....)))))))))......................((.(((((((.(((....))).((((.((.....)).))))))))))).)). (-26.45 = -28.23 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:04 2006