
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,694,700 – 5,694,793 |
| Length | 93 |
| Max. P | 0.969461 |

| Location | 5,694,700 – 5,694,793 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.66 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.66 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5694700 93 + 22224390 UACCGAAUUUCAAAAGCGACGACCACAAGCCGCCGCGCAGAACAGCACCUUCUGCUUCCAAUUC--------UCCAGCUUCUACACGU-UGAAAAA----------UU------UCUG ....(((((......(((.((.........)).)))((((((.......))))))....)))))--------..((((........))-)).....----------..------.... ( -16.00) >DroSec_CAF1 15449 94 + 1 UCCCGAAUUUCAAAAGCGGCGACCACAAGCCGCCGCGCAGAACAGCACCUUCUGCUUCCAAUUC--------GCCAGCUUCUACACGUGUAAAAAA----------UG------UUCG ...((((((......((((((.........))))))((((((.......))))))....)))))--------).((..((.(((....))).))..----------))------.... ( -22.80) >DroSim_CAF1 18484 94 + 1 UCCCGAAUUUCAAAAGCGGCGACCACAAGCCGCCGCGCAGAACAGCACCUUCUGCUUCCAAUUC--------GCCAGCUUUUACACGUGUAAAAAA----------UG------UUGG ...((((((......((((((.........))))))((((((.......))))))....)))))--------)(((((((((((....))))))..----------.)------)))) ( -29.70) >DroYak_CAF1 18205 108 + 1 UCCCGAAUUUCAAAAGCGGCGACCACAAGCCGCCGCGCAGAACAGCACCUUCUGCUCACAAUUCUGUAAUUCUGUAGCUUCUGCACUUGCAAAAAU----------CGUACACUUGCG ....(((((.((...((((((.........))))))((((((.......)))))).........)).)))))(((((...)))))...((((....----------.......)))). ( -26.30) >DroAna_CAF1 12537 89 + 1 UCCCCA-----AAAAGCGGCGACCACAAGCCGCAGUAAAGAGCAGAAUUUUCGCCUUUUCA--------------GG---CCACUCGG-AGAAAAACAACACCUCAUU------UCGA ((.((.-----....(((((........)))))......(((..((....))((((....)--------------))---)..)))))-.))................------.... ( -20.20) >consensus UCCCGAAUUUCAAAAGCGGCGACCACAAGCCGCCGCGCAGAACAGCACCUUCUGCUUCCAAUUC________GCCAGCUUCUACACGUGUAAAAAA__________UG______UCCG ...............(((((........)))))...((((((.......))))))............................................................... (-13.02 = -13.66 + 0.64)

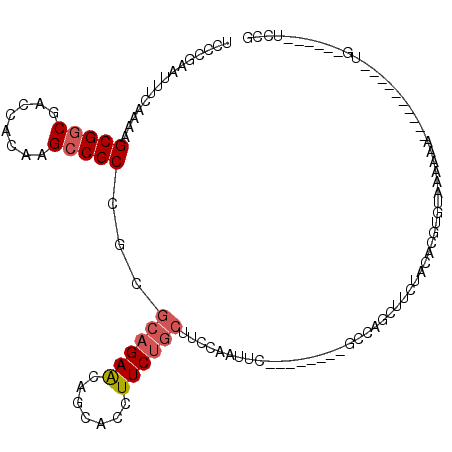
| Location | 5,694,700 – 5,694,793 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.66 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
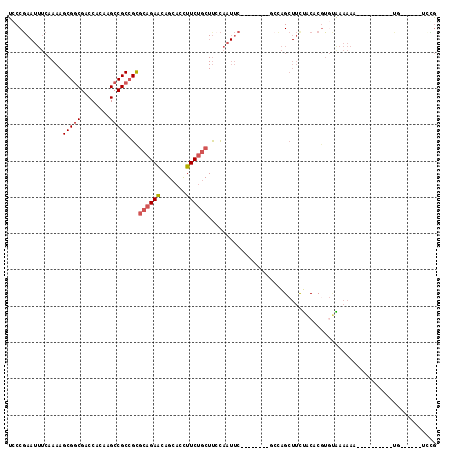
>X_DroMel_CAF1 5694700 93 - 22224390 CAGA------AA----------UUUUUCA-ACGUGUAGAAGCUGGA--------GAAUUGGAAGCAGAAGGUGCUGUUCUGCGCGGCGGCUUGUGGUCGUCGCUUUUGAAAUUCGGUA ....------..----------.......-..........((((((--------...(..(((((.....((((......))))((((((.....)))))))))))..)..)))))). ( -28.90) >DroSec_CAF1 15449 94 - 1 CGAA------CA----------UUUUUUACACGUGUAGAAGCUGGC--------GAAUUGGAAGCAGAAGGUGCUGUUCUGCGCGGCGGCUUGUGGUCGCCGCUUUUGAAAUUCGGGA ....------((----------.(((((((....))))))).)).(--------(((((....((((((.......))))))((((((((.....))))))))......))))))... ( -34.10) >DroSim_CAF1 18484 94 - 1 CCAA------CA----------UUUUUUACACGUGUAAAAGCUGGC--------GAAUUGGAAGCAGAAGGUGCUGUUCUGCGCGGCGGCUUGUGGUCGCCGCUUUUGAAAUUCGGGA ((..------((----------.(((((((....))))))).)).(--------(((((....((((((.......))))))((((((((.....))))))))......)))))))). ( -35.50) >DroYak_CAF1 18205 108 - 1 CGCAAGUGUACG----------AUUUUUGCAAGUGCAGAAGCUACAGAAUUACAGAAUUGUGAGCAGAAGGUGCUGUUCUGCGCGGCGGCUUGUGGUCGCCGCUUUUGAAAUUCGGGA (....)((((.(----------.(((((((....))))))))))))(((((.(((((..((((((((......)))))).))((((((((.....))))))))))))).))))).... ( -39.00) >DroAna_CAF1 12537 89 - 1 UCGA------AAUGAGGUGUUGUUUUUCU-CCGAGUGG---CC--------------UGAAAAGGCGAAAAUUCUGCUCUUUACUGCGGCUUGUGGUCGCCGCUUUU-----UGGGGA ..((------((..(....)..))))(((-((((((.(---((--------------(....))))((....)).))))......(((((........)))))....-----.))))) ( -26.40) >consensus CCAA______CA__________UUUUUUACACGUGUAGAAGCUGGA________GAAUUGGAAGCAGAAGGUGCUGUUCUGCGCGGCGGCUUGUGGUCGCCGCUUUUGAAAUUCGGGA ...................................(((((((.....................((((((.......))))))..((((((.....))))))))))))).......... (-19.48 = -19.80 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:01 2006