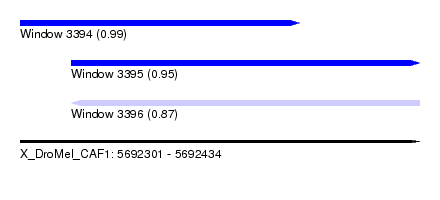
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,692,301 – 5,692,434 |
| Length | 133 |
| Max. P | 0.989306 |

| Location | 5,692,301 – 5,692,394 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 63.26 |
| Mean single sequence MFE | -18.16 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.43 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5692301 93 + 22224390 GGUACCGCAAAAAGA--------------AAAAC----AAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCGUAAUGGAAUAUGAAUAUCAAAUUCA (((...(((......--------------.....----..)))....)))...(((((..(((((((((..........))))))))))))))...(((((.....))))) ( -20.12) >DroPse_CAF1 56838 93 + 1 AGCACUGAGAGACAGAU------------AUAUCGAUGGUUGCCAUUCGCAUUCCCGUUCCACCCACUGAACUUAUACGC-UUCUCUAACGGGAUACAUGUAGCCA----- ....(((.....)))..------------.......(((((((.............((((........))))......(.-.((((....))))..)..)))))))----- ( -16.50) >DroSec_CAF1 13349 92 + 1 GGUACCGCAAAAA-A--------------AAAAC----AAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCAUAAUCGAAUAUGAAUAUCAAAUUCA (((...(((....-.--------------.....----..)))....))).........((((((((((..........))))))))))..((((.((.....)).)))). ( -18.80) >DroSim_CAF1 16155 91 + 1 GGUACCGCAAAAA----------------AAAAC----AAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCAUAAUGGAAUAUGAAUAUCAAAUUCA (((...(((....----------------.....----..)))....)))...(((((..(((((((((..........))))))))))))))...(((((.....))))) ( -20.70) >DroEre_CAF1 12708 102 + 1 GGCACCGCAAAACAAAAAACAAAAAACAAAAAAC----AA-----AAACCGCCUCCAUUUUAUGUAGUGCUAUUAAAACCACUGCGUAAUGGAAUAUGAAUAUCAAAUUCA (((...............................----..-----.....)))(((((..(((((((((..........))))))))))))))...(((((.....))))) ( -16.36) >DroPer_CAF1 60194 93 + 1 AGCACUGAGAGACAGAU------------AUAUCGAUGGUUGCCAUUCGCAUUUCCGUUCCACACAUUGAACUUAUACGC-UUCUCUAACGGGAUACAUGUAGCCA----- ....(((.....)))..------------.......(((((((.............((((........))))......(.-.((((....))))..)..)))))))----- ( -16.50) >consensus GGCACCGCAAAAAAA______________AAAAC____AAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCAUAAUGGAAUAUGAAUAUCAAAUUCA .....................................................((((((..((((((((..........)))))))))))))).................. ( -9.90 = -10.43 + 0.53)

| Location | 5,692,318 – 5,692,434 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5692318 116 + 22224390 AACAAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCGUAAUGGAAUAUGAAUAUCAAAUUCAGACUCAGUGGCUAGCAAAGUGUUUCACCUCCAGUUGGAUG ((((.(((.....((((.(((((..(((((((((..........))))))))))))))...(((((.....)))))......))))...)))...)))).....(((....))).. ( -28.20) >DroSec_CAF1 13365 113 + 1 AACAAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCAUAAUCGAAUAUGAAUAUCAAAUUCAGACUCAGUGGUUGGCAAAGUGUUUCCCCUCCAUUUGG--- ((((.(((((..(((((.......((((((((((..........))))))))))..((...(((((.....)))))...)).))))))))))...))))..............--- ( -31.20) >DroSim_CAF1 16170 113 + 1 AACAAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCAUAAUGGAAUAUGAAUAUCAAAUUCAGACUCAGUGGCUGGCAAAGUGUUUCCCCUCCAUUUGG--- ((((.(((((...((((.(((((..(((((((((..........))))))))))))))...(((((.....)))))......)))).)))))...))))..............--- ( -31.80) >DroEre_CAF1 12739 110 + 1 AACAA-----AAACCGCCUCCAUUUUAUGUAGUGCUAUUAAAACCACUGCGUAAUGGAAUAUGAAUAUCAAAUUCAGACUCAGUGGCCAGAAAAGUGUUU-CCCUCCAAUUAGAUG ((((.-----.....((((((((..(((((((((..........))))))))))))))...(((((.....)))))........)))........)))).-............... ( -20.34) >DroYak_CAF1 13675 115 + 1 AACAAUGC-AAACCCACCUCCGGUUGAUGCAGUGAAAUGAAAACCACUGCGUAAUGGAAUAUGAAUAUCAUAUGCAGACUCAGUGGCUAGCAAAGUGCUGCCCCUCCAUUUUGAUG ..((((((-.........((((....((((((((..........))))))))..))))(((((.....)))))))).....((.(((.(((.....)))))).)).....)))... ( -29.30) >consensus AACAAUGCCAAAACCACCUCCGUUUUAUGCAGUGAAAUUAAAACCACUGCGUAAUGGAAUAUGAAUAUCAAAUUCAGACUCAGUGGCUAGCAAAGUGUUUCCCCUCCAUUUGGAUG ((((.(((.....((((.(((((..(((((((((..........))))))))))))))...(((((.....)))))......))))...)))...))))................. (-20.86 = -21.74 + 0.88)


| Location | 5,692,318 – 5,692,434 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -23.64 |
| Energy contribution | -25.12 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5692318 116 - 22224390 CAUCCAACUGGAGGUGAAACACUUUGCUAGCCACUGAGUCUGAAUUUGAUAUUCAUAUUCCAUUACGCAGUGGUUUUAAUUUCACUGCAUAAAACGGAGGUGGUUUUGGCAUUGUU ..(((....)))........((..(((((((((((((((.(((((.....))))).))))......(((((((........)))))))..........))))))..)))))..)). ( -33.50) >DroSec_CAF1 13365 113 - 1 ---CCAAAUGGAGGGGAAACACUUUGCCAACCACUGAGUCUGAAUUUGAUAUUCAUAUUCGAUUAUGCAGUGGUUUUAAUUUCACUGCAUAAAACGGAGGUGGUUUUGGCAUUGUU ---((....))...(....)((..(((((((((((((((.(((((.....))))).))))..(((((((((((........)))))))))))......))))))..)))))..)). ( -40.00) >DroSim_CAF1 16170 113 - 1 ---CCAAAUGGAGGGGAAACACUUUGCCAGCCACUGAGUCUGAAUUUGAUAUUCAUAUUCCAUUAUGCAGUGGUUUUAAUUUCACUGCAUAAAACGGAGGUGGUUUUGGCAUUGUU ---((....))...(....)((..(((((((((((((((.(((((.....))))).))))..(((((((((((........)))))))))))......))))))..)))))..)). ( -39.40) >DroEre_CAF1 12739 110 - 1 CAUCUAAUUGGAGGG-AAACACUUUUCUGGCCACUGAGUCUGAAUUUGAUAUUCAUAUUCCAUUACGCAGUGGUUUUAAUAGCACUACAUAAAAUGGAGGCGGUUU-----UUGUU ..........((.((-((((........((((((((.((..((((.((.....)).))))....)).))))))))......((.((.(((...))).)))).))))-----)).)) ( -22.00) >DroYak_CAF1 13675 115 - 1 CAUCAAAAUGGAGGGGCAGCACUUUGCUAGCCACUGAGUCUGCAUAUGAUAUUCAUAUUCCAUUACGCAGUGGUUUUCAUUUCACUGCAUCAACCGGAGGUGGGUUU-GCAUUGUU ((((..((((((((((((((.....))).))).))........(((((.....)))))))))))..(((((((........)))))))..........)))).....-........ ( -31.50) >consensus CAUCCAAAUGGAGGGGAAACACUUUGCUAGCCACUGAGUCUGAAUUUGAUAUUCAUAUUCCAUUACGCAGUGGUUUUAAUUUCACUGCAUAAAACGGAGGUGGUUUUGGCAUUGUU .................((((...(((((((((((((((.(((((.....))))).))))......(((((((........)))))))..........)))))))..)))).)))) (-23.64 = -25.12 + 1.48)

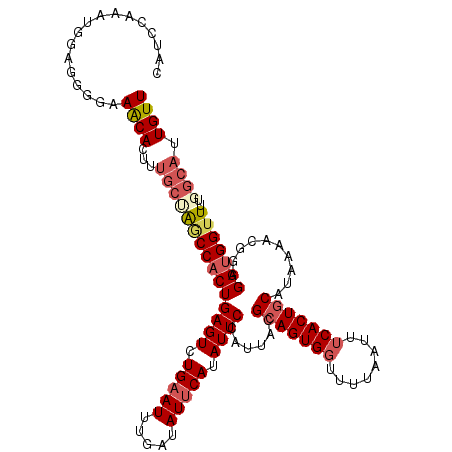
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:58 2006