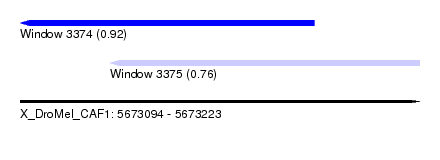
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,673,094 – 5,673,223 |
| Length | 129 |
| Max. P | 0.918792 |

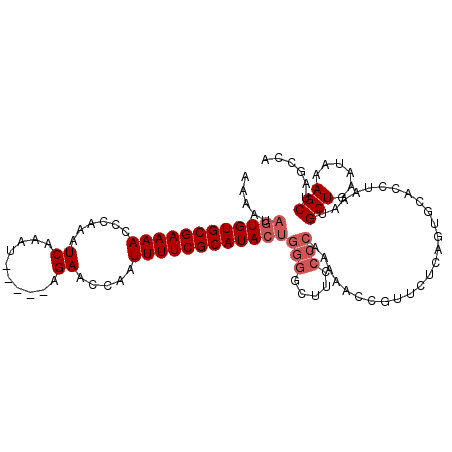
| Location | 5,673,094 – 5,673,189 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.85 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -14.17 |
| Energy contribution | -15.83 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5673094 95 - 22224390 AAAAUAAUGUGCGAAAACCCAAAUCAAAU-----AGAACCAAUUUUCGCAUACCCUAAUUCCCCCAACAA---------AUGCACCCAAAUGCUGAAUAAAGCAAGCCA .......((((((((((......((....-----.)).....))))))))))..................---------...........((((......))))..... ( -13.70) >DroSec_CAF1 12141 109 - 1 AAAAUAGUGUGCGAAAACCCAAAUCAAAUAUAGUAGAACCAAUUUUCGCAUACUGGGGCUUCCCCAAAAACCGUUCUCAGUGCACCUAAAUGCUGAAUAAAGCUAGCCA .....((((((((((((......((..........)).....))))))))))))((((...))))...........(((((..........)))))............. ( -22.60) >DroSim_CAF1 12630 104 - 1 AAAAUAGUGUGCGAAAACCCAAAUCAAAU-----AGAACCAAUUUUCGCAUACUGGGGCUUCCCCAAAAACCGUUCUCAGUGCACCUAAAUGCUGAAUAAAGCUAGCCA .....((((((((((((......((....-----.)).....))))))))))))((((...))))...........(((((..........)))))............. ( -23.50) >consensus AAAAUAGUGUGCGAAAACCCAAAUCAAAU_____AGAACCAAUUUUCGCAUACUGGGGCUUCCCCAAAAACCGUUCUCAGUGCACCUAAAUGCUGAAUAAAGCUAGCCA .....((((((((((((......((..........)).....))))))))))))(((.....)))..........................(((......)))...... (-14.17 = -15.83 + 1.67)

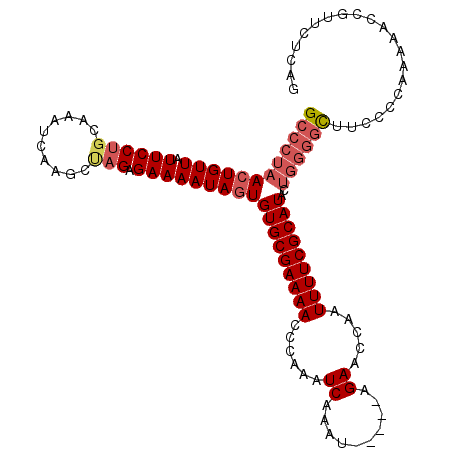
| Location | 5,673,123 – 5,673,223 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -13.06 |
| Energy contribution | -15.40 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5673123 100 - 22224390 GCCCUAAAUGUUAUUCCCUGCAAUCAAGCUAGCGAAAAUAAUGUGCGAAAACCCAAAUCAAAU-----AGAACCAAUUUUCGCAUACCCUAAUUCCCCCAACAA---------A ........((((.(((.((((......)).)).))).....((((((((((......((....-----.)).....)))))))))).............)))).---------. ( -14.50) >DroSec_CAF1 12170 114 - 1 GCCCUAACUGUUAUUCCUGCAAAUCAAGCCAGAGAAAAUAGUGUGCGAAAACCCAAAUCAAAUAUAGUAGAACCAAUUUUCGCAUACUGGGGCUUCCCCAAAAACCGUUCUCAG ((((((((((((.(((((((.......).))).)))))))))(((((((((......((..........)).....)))))))))..))))))..................... ( -25.20) >DroSim_CAF1 12659 109 - 1 GCCCUAACUGUUAUUCCUGCAAAUCAAGCUAGAGAAAAUAGUGUGCGAAAACCCAAAUCAAAU-----AGAACCAAUUUUCGCAUACUGGGGCUUCCCCAAAAACCGUUCUCAG ((((((((((((.(((((((.......)).)).)))))))))(((((((((......((....-----.)).....)))))))))..))))))..................... ( -26.10) >consensus GCCCUAACUGUUAUUCCUGCAAAUCAAGCUAGAGAAAAUAGUGUGCGAAAACCCAAAUCAAAU_____AGAACCAAUUUUCGCAUACUGGGGCUUCCCCAAAAACCGUUCUCAG ((((((((((((.((((((..........))).)))))))))(((((((((......((..........)).....)))))))))..))))))..................... (-13.06 = -15.40 + 2.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:39 2006