
| Sequence ID | X_DroMel_CAF1 |
|---|---|
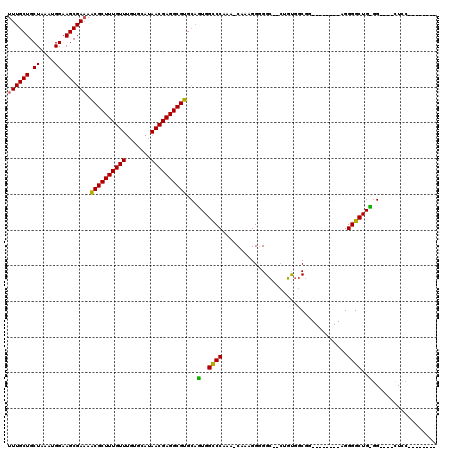
| Location | 5,670,041 – 5,670,199 |
| Length | 158 |
| Max. P | 0.931909 |

| Location | 5,670,041 – 5,670,141 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.43 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -19.23 |
| Energy contribution | -19.22 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
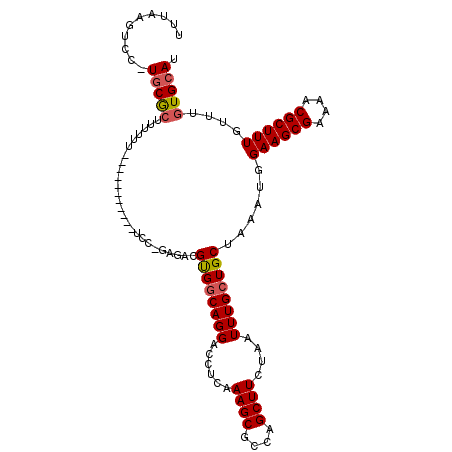
>X_DroMel_CAF1 5670041 100 - 22224390 UUUGCUGCUAAAUGGAAGCGAAAACGCUUUGUUUGUGCAUAACGAGGCGUGCAGUGGCCCAAAACAAAGGGGGCAGCUGUGGCGG--------AGGGGCUGUGG----CUCC-------- (((((((((.......)))....((((((((((.......))))))))))(((((.((((..........)))).))))))))))--------)(((((....)----))))-------- ( -38.20) >DroSec_CAF1 9067 99 - 1 UUUGCUGCUAAAUGGAAGCGAAAACGCUUUGUUUGUGCAUAACGAGGCGUGCAGUGGCCCAAA-CAAAGGAGGGGAAUCCAGCGA--------AGGGGCUGCGG----CUCC-------- ((((((.((....)).)))))).((((((((((.......))))))))))((.(..((((...-....(((......))).....--------..))))..).)----)...-------- ( -35.44) >DroAna_CAF1 13951 119 - 1 UUUGCUGCUAAAUGGAAGCGAAAGCGCUUUGUUUCUGCAUAACGAGGCGUGCACUGGUCCAAA-CAAAGGGGGCUCCAGUGGCGGCGCACUCGAGGGGCUGGCGACAGGUGCCCAGGGGC ...(((((((..((((((.(...((((((((((.......)))))))))).).)).((((...-......)))))))).)))))))((.(((...((((((....))...)))).))))) ( -45.50) >DroSim_CAF1 9603 92 - 1 UUUGCUGCUA-AUGGAAGCGA-AACGCUUUGU-UGUGCAU-ACGAGGCGUGNNNNNNNNNNNN-NNNNNNNNNNNNNNNNNNNNN--------NNNNNNNNNNN----N----------- ((((((.(..-...).)))))-)(((((((((-.......-))))))))).............-.....................--------...........----.----------- ( -15.50) >consensus UUUGCUGCUAAAUGGAAGCGAAAACGCUUUGUUUGUGCAUAACGAGGCGUGCAGUGGCCCAAA_CAAAGGGGGC__CUGUGGCGG________AGGGGCUG_GG____CUCC________ ((((((.((....)).)))))).((((((((((.......))))))))))...(..((((...................................))))..).................. (-19.23 = -19.22 + -0.00)


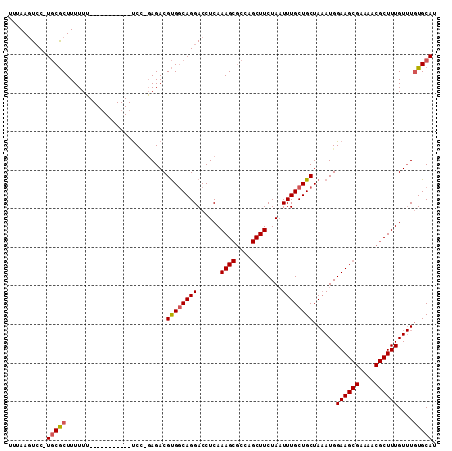
| Location | 5,670,101 – 5,670,199 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5670101 98 - 22224390 UUUAAGUCC-UGCGCUUUUUU-----------UCC-GAGACGUGGCAGGACCUCAAAGCGCCAGCUUCUAAUUUGCUGCUAAAUGGAAGCGAAAACGCUUUGUUUGUGCAU .....((((-(((((.((((.-----------...-)))).)).)))))))..(((((((...(((((((.((((....))))))))))).....)))))))......... ( -30.10) >DroSec_CAF1 9126 99 - 1 UUUAAGUCC-UGCGCUUUUUU-----------UCCAGAGACGUGGCAGGACCUCAAAGCGCCAGCUUCUAAUUUGCUGCUAAAUGGAAGCGAAAACGCUUUGUUUGUGCAU .....((((-(((((.((((.-----------....)))).)).)))))))..(((((((...(((((((.((((....))))))))))).....)))))))......... ( -30.60) >DroAna_CAF1 14030 104 - 1 UUCAACUGCUUUCACUUUUUGUGUCCUGCCGCUCU-CAGACGCGCCAGGACCUCAAAG------CUUCUAAUUUGCUGCUAAAUGGAAGCGAAAGCGCUUUGUUUCUGCAU .......((((((....((((.((((((.(((...-.....))).))))))..))))(------((((((.((((....)))))))))))))))))((.........)).. ( -34.80) >DroSim_CAF1 9658 93 - 1 UUUAAGUCC-UGCGCUUUUUU-----------CC--GAGACGUGGCAGGACCUCAAAGCGCCAGCUUCUA-UUUGCUGCUA-AUGGAAGCGA-AACGCUUUGU-UGUGCAU .....((((-(((((((((..-----------..--)))).)).)))))))..(((((((...(((((((-((.......)-))))))))..-..))))))).-....... ( -32.30) >consensus UUUAAGUCC_UGCGCUUUUUU___________UCC_GAGACGUGGCAGGACCUCAAAGCGCCAGCUUCUAAUUUGCUGCUAAAUGGAAGCGAAAACGCUUUGUUUGUGCAU ..........(((((..........................((((((((......((((....))))....))))))))......((((((....))))))....))))). (-18.26 = -18.70 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:33 2006