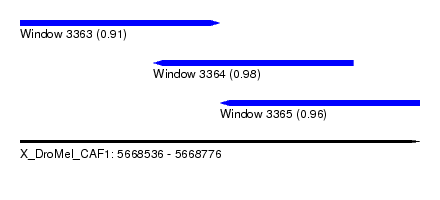
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,668,536 – 5,668,776 |
| Length | 240 |
| Max. P | 0.980290 |

| Location | 5,668,536 – 5,668,656 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -34.82 |
| Energy contribution | -35.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5668536 120 + 22224390 CCCCGACCAUUGAAAAGUUGAAAAGUGUUUGCAUUCAGCUUCAUCAGCUGAUUGCCGGCAGGUUCAAAGCAUCAAAGCGAAGAAGGCAAACUUUCAGGGGGGGGAAUGCGAAAUGGGGAA ((((..((.((((....(((((...(((..(((.((((((.....)))))).)))..)))..)))))....))))......(((((....))))).))..))))................ ( -38.20) >DroSec_CAF1 7592 117 + 1 CCCCGACCAUUGAAAUGUUGAAAAGUGUUUGCAUUCAGCUUCAUCAGCUGAUUGCCGGCAGGUUCAAAGCUUCAAAGCGAAGAAGGCAAACUUUCCGA---GGGAAUGCGAAAUGGGGAA (((((....(((((...(((((...(((..(((.((((((.....)))))).)))..)))..)))))...)))))..........(((...((((...---.)))))))....))))).. ( -35.90) >DroSim_CAF1 8395 117 + 1 CCCCGACCAUUGGAAAGUUGAAAAGUGUUUGCAUUCAGCUUCAUCAGCUGAUUGCCGGCAGGUUCAAAGCUUCAAAGCGAAGAAGGCAAACUUUCCGA---GGGAAUGCGAAAUGGGGAA ((((..((.(((((((((((((...(((..(((.((((((.....)))))).)))..)))..)))....((((........))))...))))))))))---.)).((.....)))))).. ( -41.00) >consensus CCCCGACCAUUGAAAAGUUGAAAAGUGUUUGCAUUCAGCUUCAUCAGCUGAUUGCCGGCAGGUUCAAAGCUUCAAAGCGAAGAAGGCAAACUUUCCGA___GGGAAUGCGAAAUGGGGAA (((((....(((((...(((((...(((..(((.((((((.....)))))).)))..)))..)))))...)))))..........(((...(((((.....))))))))....))))).. (-34.82 = -35.27 + 0.45)


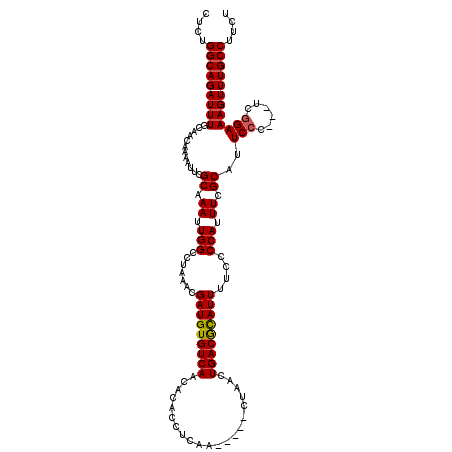
| Location | 5,668,616 – 5,668,736 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -23.51 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5668616 120 - 22224390 CUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAACUCGACUAACUGACGCAUUUUCCCCAUUUCGCAUUCCCCCCCCUGAAAGUUUGCCUUCU ....(((.(((((((..........))))))).)))((((((((((((((.....................))))))))).......(((((............))))))))))...... ( -21.80) >DroSec_CAF1 7672 112 - 1 CUCUGGCAGAUUUGCAACAAAACUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAA-----CUAACUGACAUAUUUUCCCCAUUUCGCAUUCCC---UCGGAAAGUUUGCCUUCU ....(((((((((............((.((.(((.......(((((((((...........-----.....)))))))))....))).)).))..(((.---..)))))))))))).... ( -22.89) >DroSim_CAF1 8475 112 - 1 CUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAA-----CUAACUGACGCAUUUUCCCCAUUUCGCAUUCCC---UCGGAAAGUUUGCCUUCU ....(((((((((............((.((.(((.......(((((((((...........-----.....)))))))))....))).)).))..(((.---..)))))))))))).... ( -24.39) >consensus CUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAA_____CUAACUGACGCAUUUUCCCCAUUUCGCAUUCCC___UCGGAAAGUUUGCCUUCU ....(((((((((............((.((.(((.......(((((((((.....................)))))))))....))).)).))..(((......)))))))))))).... (-23.51 = -23.40 + -0.11)


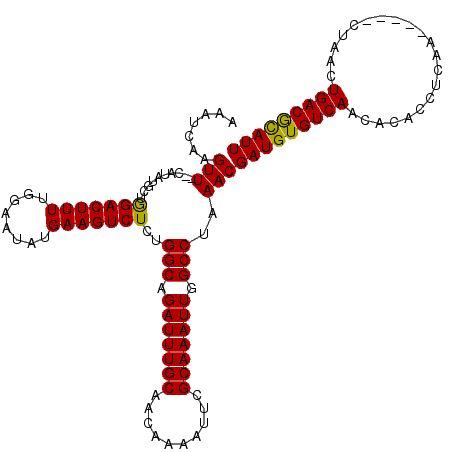
| Location | 5,668,656 – 5,668,776 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -26.03 |
| Energy contribution | -25.37 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.958912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5668656 120 - 22224390 AAAUCAAGUUCUCAUAUGCAAGACUUUUGGAAAAUGAAGUCUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAACUCGACUAACUGACGCAUU .......(((..........(((((((........)))))))..(((.(((((((..........))))))).)))..)))(((((((((.....................))))))))) ( -26.00) >DroSec_CAF1 7709 113 - 1 AAAUCAAGUU--CAUAUGCUGGACUUUUGGAAUAUGAAGUCUCUGGCAGAUUUGCAACAAAACUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAA-----CUAACUGACAUAUU ........((--(((((.(..(....)..).)))))))(((...(((.(((((((..........))))))).))).....((.((((....)))).))..-----......)))..... ( -25.40) >DroSim_CAF1 8512 113 - 1 AAAUCAAGUU--CAUAUGCUGGACUUUUGGAAUAUGAAGUCUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAA-----CUAACUGACGCAUU ........((--(((((.(..(....)..).)))))))((....(((.(((((((..........))))))).)))...))(((((((((...........-----.....))))))))) ( -26.59) >consensus AAAUCAAGUU__CAUAUGCUGGACUUUUGGAAUAUGAAGUCUCUGGCAGAUUUGCAACAAAAUUCGCAAAUUGGCCUAAACGAUGUGUCAACACACCUCAA_____CUAACUGACGCAUU .......(((..........(((((((........)))))))..(((.(((((((..........))))))).)))..)))(((((((((.....................))))))))) (-26.03 = -25.37 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:30 2006