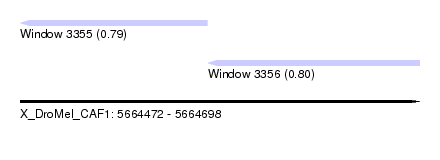
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,664,472 – 5,664,698 |
| Length | 226 |
| Max. P | 0.804987 |

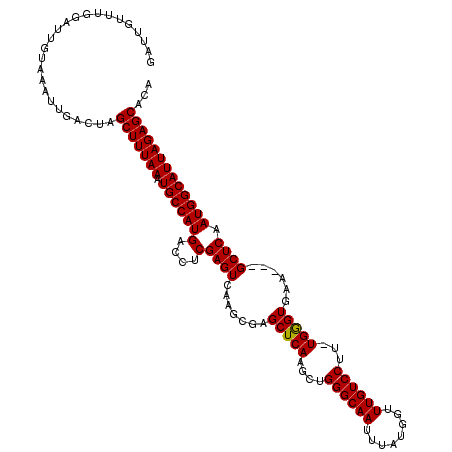
| Location | 5,664,472 – 5,664,578 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -17.35 |
| Energy contribution | -18.69 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5664472 106 - 22224390 AAGGAAAAGGACAUGUGCGACAUGGUGCAUAGUUCGGUUUUGACUAUGUGCACAGCUGCACAUAUUUAAGCGCCUUUAACAAGCACUCCAUUCGCACUUUG--------AACUU------ ......(((..((.((((((.(((((((...(((.((((((((.((((((((....)))))))).))))).)))...)))..)))..))))))))))..))--------..)))------ ( -32.60) >DroSec_CAF1 3686 108 - 1 AAGGAAAAGGACAUGUGGCACAUAGUGCAUAGUUCGGCA------------GCAGCUUCGCAUAUUUAAGCUCCUUUAACAAGCACUCUAUUCGCACUUUGUGCUAUUUAACUUGGAUUU ......(((.....((((((((.(((((((((....(((------------(.(((((.........))))).)).......))...))))..))))).))))))))....)))...... ( -26.01) >DroSim_CAF1 4047 108 - 1 AAGGAAAAGGACAUGUGGCACAAAGUGCAUAGUUCGGCA------------GCAGCUUCGCAUAUUUAAGCUCCUUUAACAAGCACUCCAUACGCACUUUGUGCUAUUUAACUUGAAGCU ......(((.....((((((((((((((...((..(((.------------...)))..))........(((.........))).........))))))))))))))....)))...... ( -29.20) >consensus AAGGAAAAGGACAUGUGGCACAUAGUGCAUAGUUCGGCA____________GCAGCUUCGCAUAUUUAAGCUCCUUUAACAAGCACUCCAUUCGCACUUUGUGCUAUUUAACUUG_A__U ......(((.....((((((((.(((((...((..(((................)))..))........((...........)).........))))).))))))))....)))...... (-17.35 = -18.69 + 1.34)

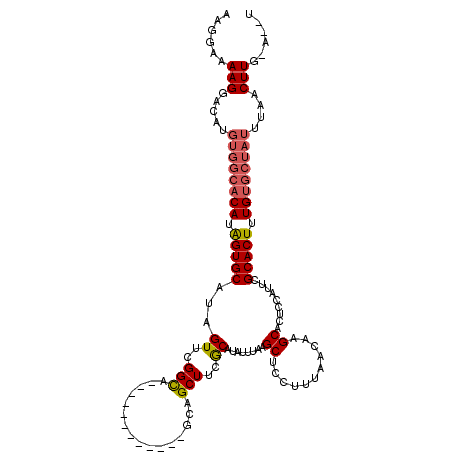
| Location | 5,664,578 – 5,664,698 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.63 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -33.07 |
| Energy contribution | -32.85 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5664578 120 - 22224390 GAUUGUUUGGAUUGUAAAUUGACUAGCUUUAAAUGCCAUGACCUCGAGUCAAGCGAGCUCAAGCUGGGCAAUUAAUGGUUUGUCCUUUUGAGUGAAGCAGCUCAAUGGCAUUAGAGCACA .........................(((((((.(((((((....)((((...((..(((((((..((((((........)))))).)))))))...)).)))).)))))))))))))... ( -37.40) >DroSec_CAF1 3794 116 - 1 GAUUGUUUGGAUUGUAAAUUGACUAGCUUUAAAUGCCAUGACCUCGAGUCAAGCGAGCUCAAGCUGGGCAAUUUAUGGUUUGUCCUU-UGGGUGAA---GCUCAAUGGCAUUAGAGCACA .........................(((((((.(((((((((.....))).(((..(((((((..((((((........))))))))-)))))...---)))..)))))))))))))... ( -35.20) >DroSim_CAF1 4155 116 - 1 GAUUGUUUGGAUUGUAAAUUGACUAGCUUUAAAUGCCAUGACCUCGAGUCAAGCGAGCUCAAGCUGGGCAAUUUAUGGUUUGUCCUU-UGGGUGAA---GCUCAAUGGCAUUAGAGCACA .........................(((((((.(((((((((.....))).(((..(((((((..((((((........))))))))-)))))...---)))..)))))))))))))... ( -35.20) >consensus GAUUGUUUGGAUUGUAAAUUGACUAGCUUUAAAUGCCAUGACCUCGAGUCAAGCGAGCUCAAGCUGGGCAAUUUAUGGUUUGUCCUU_UGGGUGAA___GCUCAAUGGCAUUAGAGCACA .........................(((((((.(((((((....)((((.......(((((....((((((........))))))...)))))......)))).)))))))))))))... (-33.07 = -32.85 + -0.22)

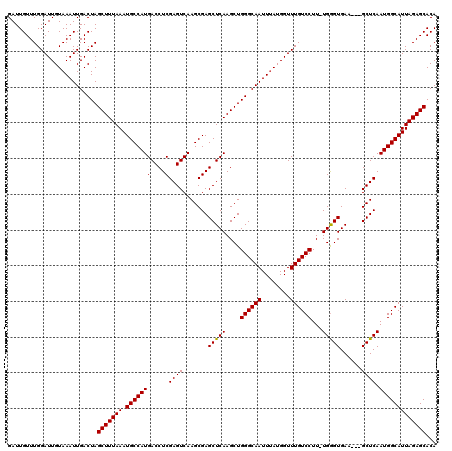
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:22 2006