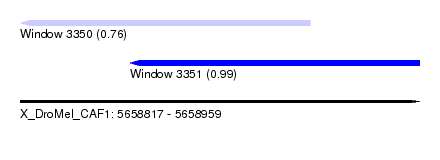
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,658,817 – 5,658,959 |
| Length | 142 |
| Max. P | 0.987794 |

| Location | 5,658,817 – 5,658,920 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.47 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -16.55 |
| Energy contribution | -17.55 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
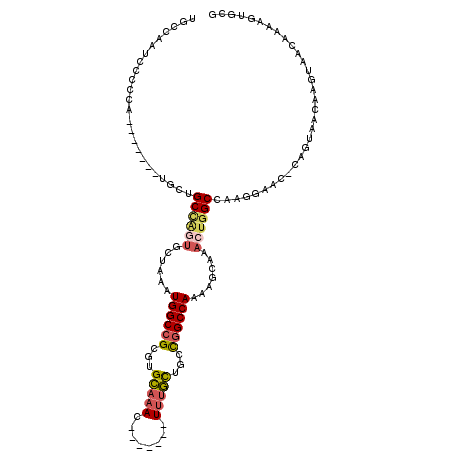
>X_DroMel_CAF1 5658817 103 - 22224390 UGCCAAUCCCCCA-------UGCUGCCAGUGCUAAAUGGCCGCGUGCAAAC-------UUUGCUGCCGGCCAAAAGCAAACUGGCCAAGGAACCCAGUAACAAGUAACAAAAGUGCG .(((......((.-------((..(((((((((...((((((.(((((...-------..))).))))))))..)))..)))))))).))......((........))....).)). ( -29.70) >DroVir_CAF1 33632 110 - 1 UGCUGCUGCUGGUUGCCACCGGCAGCGCCAGCUAAAUGGCAGCCUGAUUACCUCUUUUUUCUUUGUUCGCCAAAGACAAACAAGC-----GGCUCGGGCAAAAGUAACAAAAGUA-- (((((((((((((....)))))))))((((......)))).((((((...((.((((((((((((.....)))))).))).))).-----)).))))))...)))).........-- ( -41.30) >DroSec_CAF1 4603 102 - 1 UGGCAAUCCCCCA-------UGCUGCUAGUGCUAAAUGGCCGCGUGCAAAC-------UUUGCUGCCGGCCAAAACCAAACUGGCCAAGGAAC-CAGUAACAAGUAACAAAAGUGCG (((((........-------...)))))(..((...((((((.(((((...-------..))).)))))))).......(((((........)-)))).............))..). ( -28.20) >DroYak_CAF1 3494 102 - 1 UGCCAAUGCCCCA-------UGCUGCCGAUGCUAAAUGGCCGCGUGCAAAC-------UUUGCUGCCGGCCAAAAGCAAACUGGCCAAGGAAC-CAGUAACAAGUAACAAAAGUGCG .......(((...-------((.(((...((((...((((((.(((((...-------..))).))))))))..)))).(((((........)-)))).....))).))...).)). ( -27.30) >consensus UGCCAAUCCCCCA_______UGCUGCCAGUGCUAAAUGGCCGCGUGCAAAC_______UUUGCUGCCGGCCAAAAGCAAACUGGCCAAGGAAC_CAGUAACAAGUAACAAAAGUGCG ........................((((((......((((((...(((((........)))))...)))))).......))))))................................ (-16.55 = -17.55 + 1.00)

| Location | 5,658,856 – 5,658,959 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.99 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
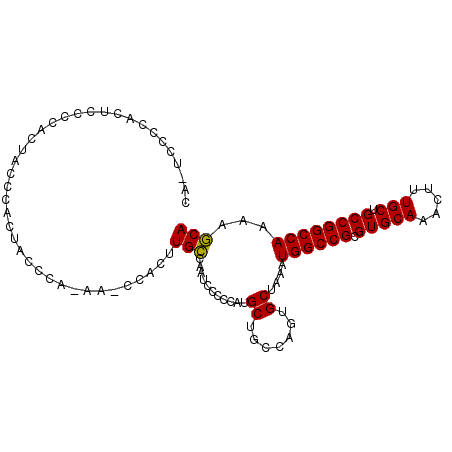
>X_DroMel_CAF1 5658856 103 - 22224390 CACUCCCCACUCCCCACUGCCCACUACCCACAAUCCACUUGCCAAUCCCCCAUGCUGCCAGUGCUAAAUGGCCGCGUGCAAACUUUGCUGCCGGCCAAAAGCA ..................((...............((((.((((........))..)).)))).....((((((.(((((.....))).))))))))...)). ( -19.80) >DroSec_CAF1 4641 98 - 1 -----CCCACUACCCACUACCCACUACCCAUAACCCACUUGGCAAUCCCCCAUGCUGCUAGUGCUAAAUGGCCGCGUGCAAACUUUGCUGCCGGCCAAAACCA -----..............................((((.((((........))))...)))).....((((((.(((((.....))).))))))))...... ( -20.90) >DroYak_CAF1 3532 89 - 1 CAUUCUCCAUUCUUCAUUCCCC--------------ACUUGCCAAUGCCCCAUGCUGCCGAUGCUAAAUGGCCGCGUGCAAACUUUGCUGCCGGCCAAAAGCA ......................--------------....((....((.....)).))...((((...((((((.(((((.....))).))))))))..)))) ( -18.20) >consensus CA_UCCCCACUCCCCACUACCCACUACCCA_AA_CCACUUGCCAAUCCCCCAUGCUGCCAGUGCUAAAUGGCCGCGUGCAAACUUUGCUGCCGGCCAAAAGCA .......................................(((...........((.......))....((((((.(((((.....))).))))))))...))) (-17.58 = -17.13 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:17 2006