
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,656,730 – 5,656,825 |
| Length | 95 |
| Max. P | 0.930547 |

| Location | 5,656,730 – 5,656,825 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.16 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
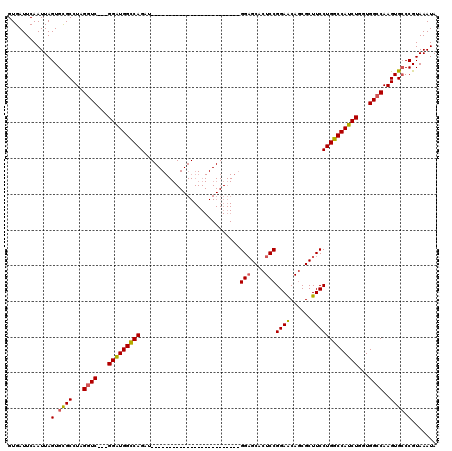
>X_DroMel_CAF1 5656730 95 + 22224390 UAUUUACGGGCACUUGGCCAACAGAUGGCCAGGAAGCGCUGUUCCGGAGUGCUCC-------------------------AUCUGGCCAUCCGCCGACCUAGGCGCACUAAUUGAAUCAC ........(((.....)))....((((((((((.((((((.......))))))..-------------------------.))))))))))((((......))))............... ( -35.30) >DroSec_CAF1 2613 92 + 1 UAUUUACGGGCACUUGGCCACCAGAUGGCCAGGAAGCGCUGUUCCGGAGUGCUCC-------------------------AUCUGGCCAUCC---GACCUAGGCGCACUAAUUGAAUCAC .......((((.(((((....(.((((((((((.((((((.......))))))..-------------------------.)))))))))).---)..))))).)).))........... ( -31.80) >DroSim_CAF1 2323 92 + 1 UAUUUACGGGCACUUGGCCACCAGAUGGCCAGGAAGCGCUGUUCCGGAGUGCUCC-------------------------AUCUGGCCAUCC---GACCUAGGCACACUAAUUGAAUCAC ......(((((.....)))....((((((((((.((((((.......))))))..-------------------------.)))))))))))---)...(((.....))).......... ( -30.30) >DroEre_CAF1 2253 92 + 1 UAUUUACGGGCACUUGGCCACCAGAUGGCCAGGAAGCGCUGCUCCGGAGCGCUCC-------------------------AGCUGGCCACCC---GAGCUAGGCGCACUAAUUGAAUCAC ......((((((((((((((.....))))))))......))).)))..(((((..-------------------------((((((....))---.)))).))))).............. ( -33.30) >DroYak_CAF1 2447 117 + 1 UAUUUACGGCCACUUGGCCACCAGAUGGCCAGGAAGCGUUGUUCCGGAGUGCACCGGAGUGGCCAUCUGGCCAUCUGGCCAUCUGACCAUCC---GACCUAGGCGCACUAAUUGAAUCAC .......((((....))))..((((((((((((........((((((......))))))(((((....)))))))))))))))))......(---((..(((.....))).)))...... ( -48.80) >consensus UAUUUACGGGCACUUGGCCACCAGAUGGCCAGGAAGCGCUGUUCCGGAGUGCUCC_________________________AUCUGGCCAUCC___GACCUAGGCGCACUAAUUGAAUCAC .......((.((((((((((.....))))))))..(((((.......)))))...............................)).))...........(((.....))).......... (-23.48 = -23.16 + -0.32)

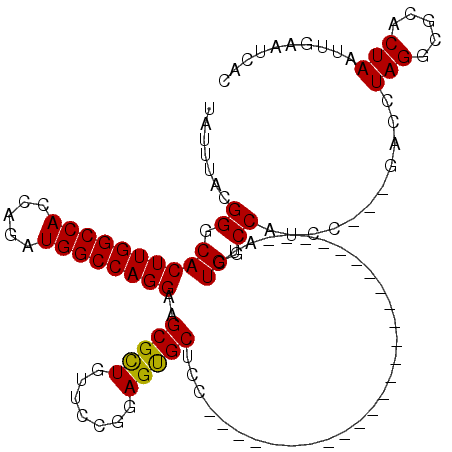

| Location | 5,656,730 – 5,656,825 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -29.24 |
| Energy contribution | -29.20 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5656730 95 - 22224390 GUGAUUCAAUUAGUGCGCCUAGGUCGGCGGAUGGCCAGAU-------------------------GGAGCACUCCGGAACAGCGCUUCCUGGCCAUCUGUUGGCCAAGUGCCCGUAAAUA ............(.((((...(((((((((((((((((..-------------------------(((((.((.......)).))))))))))))))))))))))..)))).)....... ( -43.60) >DroSec_CAF1 2613 92 - 1 GUGAUUCAAUUAGUGCGCCUAGGUC---GGAUGGCCAGAU-------------------------GGAGCACUCCGGAACAGCGCUUCCUGGCCAUCUGGUGGCCAAGUGCCCGUAAAUA ............(.((((...((((---((((((((((..-------------------------(((((.((.......)).)))))))))))))))...))))..)))).)....... ( -34.10) >DroSim_CAF1 2323 92 - 1 GUGAUUCAAUUAGUGUGCCUAGGUC---GGAUGGCCAGAU-------------------------GGAGCACUCCGGAACAGCGCUUCCUGGCCAUCUGGUGGCCAAGUGCCCGUAAAUA ........(((..((.((((.((((---((((((((((..-------------------------(((((.((.......)).)))))))))))))))...)))).)).)).))..))). ( -33.70) >DroEre_CAF1 2253 92 - 1 GUGAUUCAAUUAGUGCGCCUAGCUC---GGGUGGCCAGCU-------------------------GGAGCGCUCCGGAGCAGCGCUUCCUGGCCAUCUGGUGGCCAAGUGCCCGUAAAUA ............(.((((...((((---((((((((((..-------------------------((((((((.......)))))))))))))))))))..)))...)))).)....... ( -39.20) >DroYak_CAF1 2447 117 - 1 GUGAUUCAAUUAGUGCGCCUAGGUC---GGAUGGUCAGAUGGCCAGAUGGCCAGAUGGCCACUCCGGUGCACUCCGGAACAACGCUUCCUGGCCAUCUGGUGGCCAAGUGGCCGUAAAUA ((..(((....((((((((......---(((((((((..(((((....)))))..)))))).)))))))))))...)))..))(((.(.(((((((...))))))).).)))........ ( -52.30) >consensus GUGAUUCAAUUAGUGCGCCUAGGUC___GGAUGGCCAGAU_________________________GGAGCACUCCGGAACAGCGCUUCCUGGCCAUCUGGUGGCCAAGUGCCCGUAAAUA ............(.((((...((((...(((((((((............................(((....)))((((......)))))))))))))...))))..)))).)....... (-29.24 = -29.20 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:15 2006