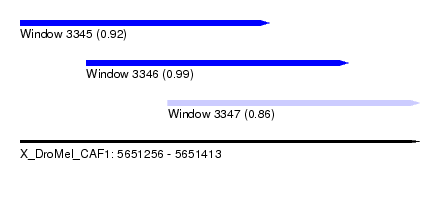
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,651,256 – 5,651,413 |
| Length | 157 |
| Max. P | 0.992304 |

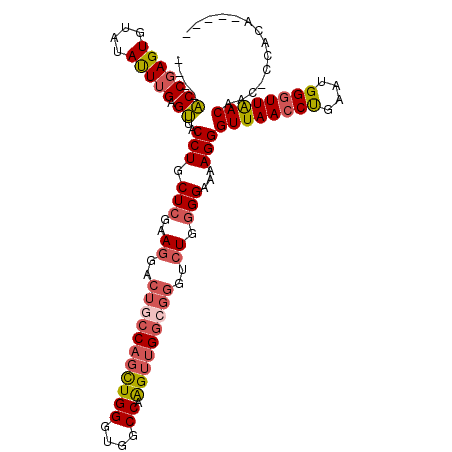
| Location | 5,651,256 – 5,651,354 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -33.29 |
| Energy contribution | -33.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5651256 98 + 22224390 UUAUUGUUCCUUCCCGUUAUGCAUUU--ACCGAGUGUAUAUUUGAGUUCCCUGCUCGAAGGACUGCCACUUGGGUGGCCAAGUUGGCGGGUCUGGGGAAA ..........(((((..(((((((((--...)))))))))(((((((.....)))))))((((((((((((((....))))).)))).))))).))))). ( -37.10) >DroSec_CAF1 40585 100 + 1 UUAUUGUUCCUUCCCGUUAUGCAUUUGCACCGAGUUUAUAUUUGAGUUACCUGCUCGAAGGACUGCCAGUUGGGUGGCCAAGUUGGCGGGUCUGGGGAAA ......(((((.((((...(((....)))((((.((....(((((((.....)))))))((.(..((.....))..))))).))))))))...))))).. ( -32.10) >DroSim_CAF1 39665 100 + 1 UUAUUGUUCCUUCCCGUUAUGCAUUUGCACCGAGUGUAUAUUUGAGUUACCUGCUCGAAGGACUACCAGCUGGGUGGCCAAGUUGGCGGGUCUGGGGAAA ......(((((.(((((((((((((((...))))))))))(((((((.....)))))))((.(((((.....)))))))......)))))...))))).. ( -36.10) >consensus UUAUUGUUCCUUCCCGUUAUGCAUUUGCACCGAGUGUAUAUUUGAGUUACCUGCUCGAAGGACUGCCAGUUGGGUGGCCAAGUUGGCGGGUCUGGGGAAA ......(((((.((((((((((((((.....)))))))))(((((((.....)))))))((.(((((.....)))))))......)))))...))))).. (-33.29 = -33.40 + 0.11)

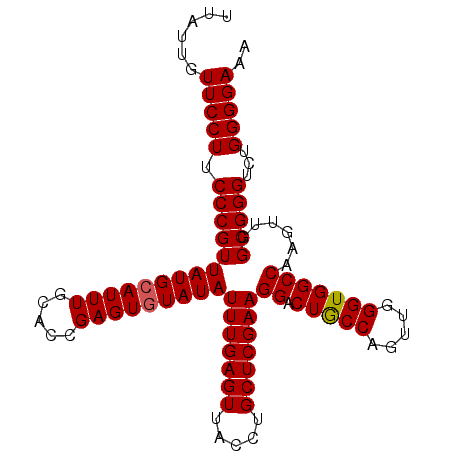

| Location | 5,651,282 – 5,651,385 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.38 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -24.96 |
| Energy contribution | -26.40 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5651282 103 + 22224390 ----ACCGAGUGUAUAUUUGAGUUCCCUGCUCGAAGGACUGCCACUUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCCGAAUGGGUUAACAAC-UCACA----- ----..............((((((.(((.(((..((..(((((((((((....))))).))))))..)).)))...)))((((((((....)))))))))))-)))..----- ( -42.80) >DroSec_CAF1 40613 104 + 1 ----ACCGAGUUUAUAUUUGAGUUACCUGCUCGAAGGACUGCCAGUUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCUGAAUGGGUUAACAGCCCCACA----- ----............(((((((.....)))))))((((((((((((((....)))).))))).)))))((((......((((((((....))))))))..))))...----- ( -39.90) >DroSim_CAF1 39693 103 + 1 ----ACCGAGUGUAUAUUUGAGUUACCUGCUCGAAGGACUACCAGCUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCUGAGUGGGUUAACAAC-CCACA----- ----.((.........(((((((.....)))))))(((((.((((((((....)).))))))..))))))).....(((.....)))..((((((.....))-)))).----- ( -35.90) >DroEre_CAF1 40696 101 + 1 AUUUGCCGAGUGUGCACUGGAGCUGCCUGCUCGA-GGACUGCCUGCUGGGUAACCAGGUUG----------GGAACAGGGUUAACCUGCUUGG-UUGACUCCCUCGCAGCAGG ....(((.(((....))).).))..(((((((((-((....(((.((((....))))...)----------))....(((((((((.....))-)))))))))))).)))))) ( -43.30) >consensus ____ACCGAGUGUAUAUUUGAGUUACCUGCUCGAAGGACUGCCAGCUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCUGAAUGGGUUAACAAC_CCACA_____ ....(((((((....))))).))..(((.(((..((..(((((((((((....)).)))))))))..)).)))...)))((((((((....)))))))).............. (-24.96 = -26.40 + 1.44)


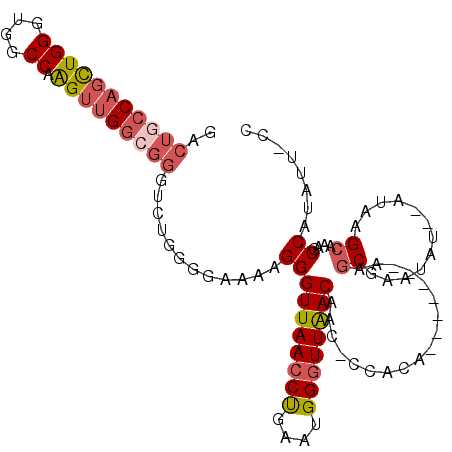
| Location | 5,651,314 – 5,651,413 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.54 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -17.79 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5651314 99 + 22224390 GACUGCCACUUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCCGAAUGGGUUAACAAC-UCACA-------UAGCGAUUA---GUAAGCAAAGCCAUAUU-CC (((((((((((((....))))).)))).))))...((((..((((((((((....))))))))...-.....-------..((.....---....))....))...))-)) ( -33.10) >DroSec_CAF1 40645 101 + 1 GACUGCCAGUUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCUGAAUGGGUUAACAGCCCCACA-------UAGCGAAUAU--AUAAGCAAAGCCAUAUU-CC (((((((((((((....)))).))))).))))(((((......((((((((....))))))))..)))))..-------....((((((--....((...)).)))))-). ( -34.20) >DroSim_CAF1 39725 100 + 1 GACUACCAGCUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCUGAGUGGGUUAACAAC-CCACA-------AAGCGAAUAU--AUAAGCAAAGCCAUAUU-CC ((((.((((((((....)).))))))..))))...((((...((((.......((((((.....))-)))).-------..((......--....))..))))...))-)) ( -31.50) >DroEre_CAF1 40731 100 + 1 GACUGCCUGCUGGGUAACCAGGUUG----------GGAACAGGGUUAACCUGCUUGG-UUGACUCCCUCGCAGCAGGCAAUUCGAAUCGAGCCCGGCAAAGCCGUAUCUAA ...(((((((((.((..((.....)----------)..)).(((((((((.....))-))))))).....))))))))).........((...((((...))))..))... ( -38.50) >consensus GACUGCCAGCUGGGUGGCCAAGUUGGCGGGUCUGGGGAAAAGGGUUAACCUGAAUGGGUUAACAAC_CCACA_______AAGCGAAUAU__AUAAGCAAAGCCAUAUU_CC ..(((((((((((....)).)))))))))............((((((((((....))))))))..................((............))....))........ (-17.79 = -19.22 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:14 2006