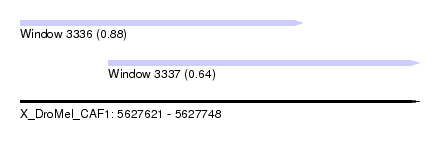
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,627,621 – 5,627,748 |
| Length | 127 |
| Max. P | 0.878844 |

| Location | 5,627,621 – 5,627,711 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -16.55 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
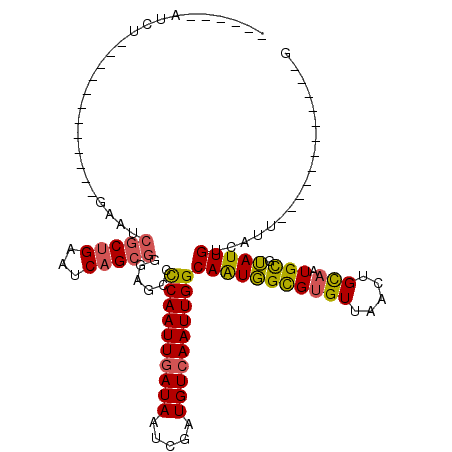
>X_DroMel_CAF1 5627621 90 + 22224390 CUCGAGCUCCAUUUCCAUACC------GAUCUCG------------AUCU------------GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGUGUG ...............((((((------(.(((((------------((..------------..)))((((...))))))))...((((((((((.....))))))))))...))))))) ( -26.30) >DroSec_CAF1 17276 90 + 1 CUCGAGCUCCAUUUCCAUAUC------GAUCUCG------------AUCU------------GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGUGUG .....(((((.....((.(((------(....))------------)).)------------)...(((((...)))))))))).((((((((((.....)))))))))).......... ( -29.80) >DroSim_CAF1 17621 90 + 1 CUCGAGCUCCAUUUCCAUAUC------GAUCUCG------------AUCU------------GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGUGUG .....(((((.....((.(((------(....))------------)).)------------)...(((((...)))))))))).((((((((((.....)))))))))).......... ( -29.80) >DroEre_CAF1 17448 98 + 1 CUGAGGCUCCAUUUCCAUAUC------GAUCUCG------------AUCUGAAUCG----CUGAAGCGCUGAAUCAGCGGGGCCCCCAAUUGAUAAUCGAUGUCAAUUGGCAGUGGCGCG (((.((((((.....((.(((------(....))------------)).))....(----((((.........))))))))))).((((((((((.....)))))))))))))....... ( -34.20) >DroYak_CAF1 17176 114 + 1 CUGAAGCUCCAUUUCCAUAUC------GAUCUCGAUCUCGAUCUCGAUCUCAAUCUCGAUCUGAAUCGCUGAAUCAGCGGGGGCCCCAAUUGAUAAACGAUGUCAAUUGGCAAUGGCGUG .....(((((.....((.(((------((....((..(((....)))..))....))))).))...(((((...)))))))))).((((((((((.....)))))))))).......... ( -33.30) >DroPer_CAF1 64704 100 + 1 CUGAAUCUCCCUCUCCCACUCCGUCGCAAUCUUC------------AACUGAAUCU------GAAU--CUGAAUCUGAUUCAGCCUCAAUUUAUAAUCGAUGUCAAUUGGCAAUUGCGUG ........................((((((....------------...(((..((------((((--(.......)))))))..)))............((((....)))))))))).. ( -19.00) >consensus CUCAAGCUCCAUUUCCAUAUC______GAUCUCG____________AUCU____________GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGCGUG .....((.(((((.....................................................(((((...)))))......((((((((((.....)))))))))).))))).)). (-16.55 = -17.50 + 0.95)


| Location | 5,627,649 – 5,627,748 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.55 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.15 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5627649 99 + 22224390 ------AUCU------------GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGUGUGUUAAUUGCAAUGCGUAUUGUCAUUGAAAUCGGCGUUC ------....------------(((.((((((....((....))..((((.((((((((.((.(((((((((......)))))))))))...)).))))))))))...))))))))) ( -28.90) >DroSec_CAF1 17304 84 + 1 ------AUCU------------GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGUGUGUUAAUUGCAAUGCGUAUUGUCA--------------- ------....------------.....((((...))))(((...)))...(((((((((.((.(((((((((......)))))))))))...)).)))))))--------------- ( -21.80) >DroSim_CAF1 17649 86 + 1 ------AUCU------------GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGUGUGUUAAUUGCAAUGCGUAUUGUCAUU------------- ------....------------.....((((...))))(((...)))...(((((((((.((.(((((((((......)))))))))))...)).)))))))..------------- ( -22.40) >DroEre_CAF1 17476 95 + 1 ------AUCUGAAUCG----CUGAAGCGCUGAAUCAGCGGGGCCCCCAAUUGAUAAUCGAUGUCAAUUGGCAGUGGCGCGUUAACUGUCAUGUGUAUUGUCAUU------------G ------...(((..((----(((...(((((...)))))......((((((((((.....)))))))))))))))((((((........))))))....)))..------------. ( -27.50) >DroYak_CAF1 17210 117 + 1 AUCUCGAUCUCAAUCUCGAUCUGAAUCGCUGAAUCAGCGGGGGCCCCAAUUGAUAAACGAUGUCAAUUGGCAAUGGCGUGUUAACUGCAAUGCGUAUUGUCAUUGCAAUCGUUUUUG ..(((((((........)))).....(((((...))))))))(((((((((((((.....))))))))))....)))........((((((((.....).))))))).......... ( -32.90) >DroPer_CAF1 64738 103 + 1 ------AACUGAAUCU------GAAU--CUGAAUCUGAUUCAGCCUCAAUUUAUAAUCGAUGUCAAUUGGCAAUUGCGUGUUAACUGCAUUGUUGAUUGUAGCUGGAAUUGUUACUG ------...(((..((------((((--(.......)))))))..)))................(((..(((((.((((....)).)))))))..)))(((((.......))))).. ( -24.40) >consensus ______AUCU____________GAAUCGCUGAAUCAGCGGGAGCCCCAAUUGAUAAUCGAUGUCAAUUGGCAAUGGCGUGUUAACUGCAAUGCGUAUUGUCAUU____________G ..........................(((((...)))))......((((((((((.....))))))))))(((((((((((.....))).))).))))).................. (-16.48 = -16.15 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:04 2006