
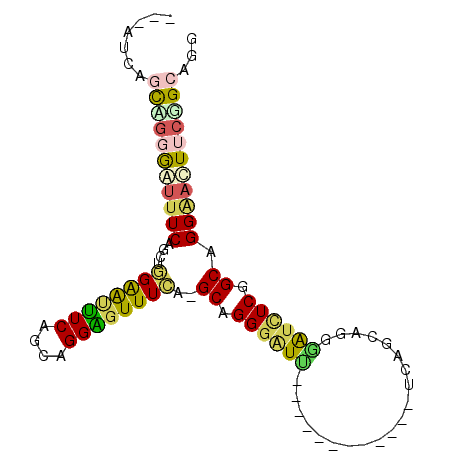
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,625,012 – 5,625,123 |
| Length | 111 |
| Max. P | 0.997213 |

| Location | 5,625,012 – 5,625,105 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 65.43 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -16.14 |
| Energy contribution | -15.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.63 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5625012 93 + 22224390 ---AUCAGCAGGGAUUUCAGCAUGGGUUUCAGCAGGAGUUUCA-GCAGGGAUCUCAGCAGGGAUUUCAGCAGGGAUCUCGGCAGGAACUUCGGCAGC ---....((.(..((((......))))..).((.((((((((.-((.((((((((.((..........)).)))))))).)).)))))))).)).)) ( -31.30) >DroSec_CAF1 14719 81 + 1 ---AUCAGCUGGGAUUUCAUCUGGGAUUUCAGCAGGGGUUUCA-GCAGGGUUU------------UCAGCAGGGAUCUCGGCAGGAACUUCGGCAGG ---....((((..((..(....)..))..)))).((((((((.-((.(((.(.------------.(....)..).))).)).))))))))...... ( -29.60) >DroSim_CAF1 15069 81 + 1 ---AUCAGCUGGGAUUUCAGCGGGGAUCUCAGCAGGGCUUUCA-GCAGGGAUC------------UCGGCUGGGAUCUCGGCAGGGAUUUCAGCAGG ---....((((..((..(.((.(((((((((((.(((.(..(.-...)..).)------------)).))))))))))).)).)..))..))))... ( -43.50) >DroYak_CAF1 14482 96 + 1 AGAAUCAGCAGGGAGUUCAGCUGGAAUUUCCAUUGGAGUUUCA-GCAGGGGUUUCAACAGGAGUUUCAGCUGGGGUUUCAGCUGGGAUUUCAGCUGG .....((((.(..(.(((.(((((((((((....)))))))))-)).))).)..)....(((((..(((((((....)))))))..))))).)))). ( -42.20) >DroMoj_CAF1 16737 76 + 1 --------CAGCAAUUUCGACUAGAGUUUCAGCUGGAGUUUUA-GCGGGAAU------------CUCAGCCUGGAUCUCUGCUGGAAUUUCUGCAGC --------(((.(((((((..(((((.((((((((..(((((.-...)))))------------..))).))))).))))).))))))).))).... ( -24.50) >DroAna_CAF1 13988 84 + 1 ---AUCUUUUUCAGUGUCGGCGGGAGCUUCGGCGGGAGUUUCUGGCUGGGAUUC----------CUCAGCAGGAGUCUCUGCUGGGACUUCGGCAAG ---...........((((..((((((((((....))))))))))((((((....----------)))))).((((((((....)))))))))))).. ( -35.60) >consensus ___AUCAGCAGGGAUUUCAGCUGGAAUUUCAGCAGGAGUUUCA_GCAGGGAUU____________UCAGCAGGGAUCUCGGCAGGAACUUCGGCAGG .......(((((((((((....((((((((....))))))))..((.((((((....................)))))).)).)))))))))))... (-16.14 = -15.98 + -0.16)


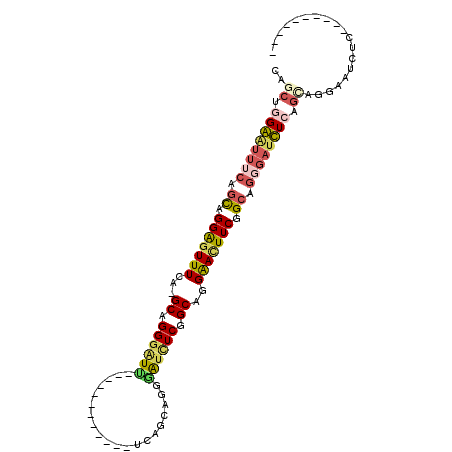
| Location | 5,625,026 – 5,625,123 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 65.42 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.55 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5625026 97 + 22224390 CAGCAUGGGUUUCAGCAGGAGUUUCA-GCAGGGAUCUCAGCAGGGAUUUCAGCAGGGAUCUCGGCAGGAACUUCGGCAGCGAUCUCAGCAGGACUCUC---------- ......((((((..((.((((((((.-((.((((((((.((..........)).)))))))).)).)))))))).)).((.......)).))))))..---------- ( -33.70) >DroSec_CAF1 14733 85 + 1 CAUCUGGGAUUUCAGCAGGGGUUUCA-GCAGGGUUU------------UCAGCAGGGAUCUCGGCAGGAACUUCGGCAGGGACCUCAGCAGGAAUCUC---------- .....((((((((.((.(((((..(.-((.(((.((------------((.((.((....)).)).)))).))).)).)..))))).)).))))))))---------- ( -29.40) >DroSim_CAF1 15083 85 + 1 CAGCGGGGAUCUCAGCAGGGCUUUCA-GCAGGGAUC------------UCGGCUGGGAUCUCGGCAGGGAUUUCAGCAGGAAUCUCGGCAGGGAUCUC---------- ((((.((((((((.((..........-)).))))))------------)).))))(((((((.((.((((((((....)))))))).)).))))))).---------- ( -41.60) >DroYak_CAF1 14499 97 + 1 CAGCUGGAAUUUCCAUUGGAGUUUCA-GCAGGGGUUUCAACAGGAGUUUCAGCUGGGGUUUCAGCUGGGAUUUCAGCUGGGAUUUCAGCAGGGGUUUC---------- ..(((((((((((....)))))))))-)).((..((((....))))..)).((((..((..(((((((....)))))))..))..)))).........---------- ( -42.60) >DroMoj_CAF1 16746 95 + 1 CGACUAGAGUUUCAGCUGGAGUUUUA-GCGGGAAU------------CUCAGCCUGGAUCUCUGCUGGAAUUUCUGCAGCGAUUUCAGCUGGAAUCUCAGCUUGGAUU (((((.(((((((((((((((((...-((((((((------------..((((..((....))))))..))))))))...)))))))))))))).))))).))).... ( -40.30) >DroAna_CAF1 14002 88 + 1 CGGCGGGAGCUUCGGCGGGAGUUUCUGGCUGGGAUUC----------CUCAGCAGGAGUCUCUGCUGGGACUUCGGCAAGGAUUUCAUUGGUGGCUUC---------- ...((((((((((....))))))))))((((((....----------)))))).((((((..((((((....))))))..))))))............---------- ( -35.40) >consensus CAGCUGGAAUUUCAGCAGGAGUUUCA_GCAGGGAUU____________UCAGCAGGGAUCUCGGCAGGAACUUCGGCAGGGAUCUCAGCAGGAAUCUC__________ ..((.((((((((.((.(((((((...((.((((((....................)))))).))..))))))).)).)))))))).))................... (-16.13 = -16.55 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:03 2006