
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,609,701 – 5,609,834 |
| Length | 133 |
| Max. P | 0.537710 |

| Location | 5,609,701 – 5,609,795 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -17.46 |
| Energy contribution | -19.47 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609701 94 - 22224390 GUGAUGGGCGAACGGCGAUAAGCAGCU------------CGACAAAAUAUUCGAUAGUCAAACAAUUUUAUUUUAUGUUUUGUUUAUGUUACAUUAAGUAACGUCA .((((((((.....((.....)).)))------------)((((((((((..(((((..........)))))..))))))))))...(((((.....))))))))) ( -20.90) >DroSec_CAF1 11366 106 - 1 GAGAUGGGCGAACGCCAAUAGGCAGUUCGCUCGAUAGAUCGACAGAAUAUUCGAUAGUCACACAAUUUUAUUUUAUGUUUUGUUUAUGUUACAUUAUGAUACGUCU .(((((((((((((((....))).))))))))((((....((((((((((..(((((..........)))))..))))))))))..))))............)))) ( -29.30) >DroSim_CAF1 13084 106 - 1 GAGAUGGGCGAACGCCAAUAGGCAGUUCGCUCGAUAGAUCGACAGAAUACUCGAUAGUCACACAAUUUUAUUUUAUGUUUUGUUUAUGUUACCUUAUGAUACGUCU .(((((((((((((((....))).))))))))((((....(((((((((...(((((..........)))))...)))))))))..))))............)))) ( -26.70) >consensus GAGAUGGGCGAACGCCAAUAGGCAGUUCGCUCGAUAGAUCGACAGAAUAUUCGAUAGUCACACAAUUUUAUUUUAUGUUUUGUUUAUGUUACAUUAUGAUACGUCU .(((((((((((((((....))).))))))).........((((((((((..(((((..........)))))..)))))))))).................))))) (-17.46 = -19.47 + 2.00)

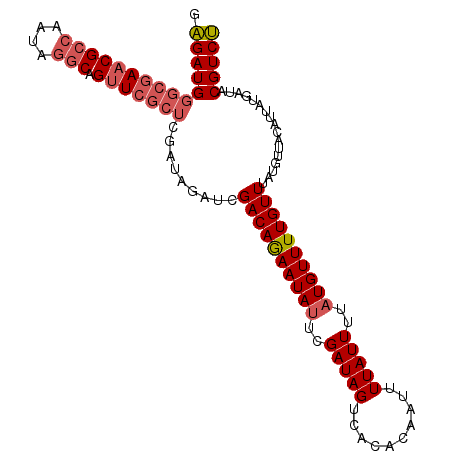

| Location | 5,609,741 – 5,609,834 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -19.22 |
| Energy contribution | -20.22 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609741 93 + 22224390 AU--U------------GUUUGACUAUCGAAUAUUUUGUCG------------AGCUGCUUAUCGCCGUUCGCCCAUCACUACUACCGACUGUCUACAAAGUGGAGUGG-GGUGUGGGUA ..--(------------(((((.....)))))).......(------------(((.((.....)).))))(((((.((((.(((((.(((........))).).))))-))))))))). ( -29.20) >DroSec_CAF1 11406 105 + 1 AU--U------------GUGUGACUAUCGAAUAUUCUGUCGAUCUAUCGAGCGAACUGCCUAUUGGCGUUCGCCCAUCUCUACUACCCACUGUCUACAAAGUGGAGUGG-GGUGUGGGUA ..--.------------((.(((..(((((........)))))...))).))((((.(((....)))))))(((((((((((((..(((((........))))))))))-)).)))))). ( -38.80) >DroSim_CAF1 13124 105 + 1 AU--U------------GUGUGACUAUCGAGUAUUCUGUCGAUCUAUCGAGCGAACUGCCUAUUGGCGUUCGCCCAUCUCUACUACCCACUGUCUACAAAGUGGAGUGG-GGUGUGGGUA ..--.------------((.(((..(((((........)))))...))).))((((.(((....)))))))(((((((((((((..(((((........))))))))))-)).)))))). ( -38.70) >DroEre_CAF1 11804 120 + 1 GUAUUUGCCUACUAGUGUCGUGACUAUCGAGUAUUUUCCCGAGCUAUCGAACCAGCUGCCUAUCGCCGUUCGCCCAUCUCUACUACCCACUGUCUAUAAAGUAGUGUGGGGGUGUGGGUA .....(((((((((((......))))...........((((((....(((((..((........)).))))).....))).....(((((((.((....))))))).))))).))))))) ( -29.60) >DroYak_CAF1 11966 116 + 1 GUAUUU---UAUUACUCCCAUGACUAUCGAAUACUUUCACGAGCUAUCAAGCCAGGCGCCUAUCGCCGUUCGCCCAUCUCUACUACCCACUGUCUACAAAGUGGUGUGG-GGUGUGGGUA (((...---...))).............((((........(.(((....)))).((((.....))))))))((((((((((((...(((((........))))).))))-)).)))))). ( -34.00) >consensus AU__U____________GUGUGACUAUCGAAUAUUUUGUCGAGCUAUCGAGCCAGCUGCCUAUCGCCGUUCGCCCAUCUCUACUACCCACUGUCUACAAAGUGGAGUGG_GGUGUGGGUA ..........................((((........)))).............................((((((..((.(((((((((........))))).)))).)).)))))). (-19.22 = -20.22 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:57 2006