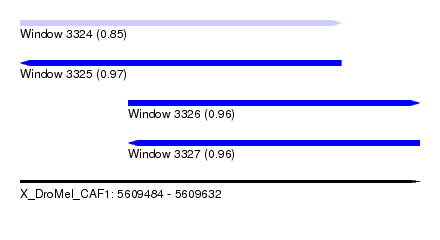
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,609,484 – 5,609,632 |
| Length | 148 |
| Max. P | 0.968477 |

| Location | 5,609,484 – 5,609,603 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -20.70 |
| Energy contribution | -24.20 |
| Covariance contribution | 3.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609484 119 + 22224390 GUUAAUCGAUUUGCGGGUUAUCGGCUUGUUCGAAAGCUAAAAAUCGAUUAAAUUGAUCGAAUACAGGAUAACGGUACGUUUUUGUAUUAGAUCUAACUGAAACUACGUUUUCCAAUUAG .(((((((((((...((((.((((.....)))).))))..)))))))))))...((((.(((((((((..(((...)))))))))))).)))).......................... ( -28.80) >DroSec_CAF1 11158 119 + 1 GUCAAUCGAUUAGCGGGUUAUCGGCUUGUACGAUAGCUAAACAUCGAUUGGAUUAAUCGAAUAAAGGAUAACGGUAUGUUUUUGUAUUCGAUCUAACUGAAAUUGCGUGUUCCAAUUAG .(((((((((.....((((((((.......))))))))....)))))))))....(((((((((((((((......))))))).))))))))((((.((.(((.....))).)).)))) ( -30.50) >DroSim_CAF1 12876 119 + 1 GUCAAUCGAUUAGCGGGUUAUCGGCUUGUACGAUAGCUAAACAUCGAUUGGAUUGAUCGAAUACAGGAUAACGGUAUGUUUUUGUAUUCGAUCUAACUGAAAUUGCGUUUUCCAAUAAG .(((((((((.....((((((((.......))))))))....)))))))))...((((((((((((((...........)))))))))))))).....((((......))))....... ( -37.20) >DroYak_CAF1 11736 104 + 1 GUCA-------------UAA--GGCUUGUUCGAUAUUUAAAAAUCGAUUAGAUUGAUCGAGUGCACAAUAACGGUAAGGUUUUGUAUUCGAUCUUACUCAAAUAGAGUUUUCCAAUUGG ....-------------...--((((...(((((........)))))..))...(((((((((((....(((......))).)))))))))))..((((.....))))...))...... ( -22.20) >consensus GUCAAUCGAUUAGCGGGUUAUCGGCUUGUACGAUAGCUAAAAAUCGAUUAGAUUGAUCGAAUACAGGAUAACGGUAUGUUUUUGUAUUCGAUCUAACUGAAAUUGCGUUUUCCAAUUAG .(((((((((.....((((((((.......))))))))....)))))))))...((((((((((((((...........)))))))))))))).......................... (-20.70 = -24.20 + 3.50)

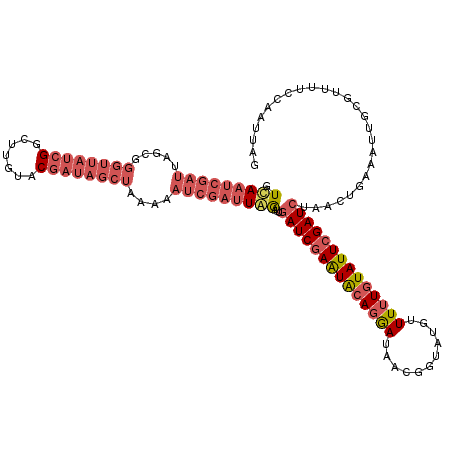
| Location | 5,609,484 – 5,609,603 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -14.56 |
| Energy contribution | -18.69 |
| Covariance contribution | 4.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609484 119 - 22224390 CUAAUUGGAAAACGUAGUUUCAGUUAGAUCUAAUACAAAAACGUACCGUUAUCCUGUAUUCGAUCAAUUUAAUCGAUUUUUAGCUUUCGAACAAGCCGAUAACCCGCAAAUCGAUUAAC .(((((((((.......)))))))))((((.((((((..((((...))))....)))))).))))...(((((((((((...((((......))))((......)).))))))))))). ( -25.60) >DroSec_CAF1 11158 119 - 1 CUAAUUGGAACACGCAAUUUCAGUUAGAUCGAAUACAAAAACAUACCGUUAUCCUUUAUUCGAUUAAUCCAAUCGAUGUUUAGCUAUCGUACAAGCCGAUAACCCGCUAAUCGAUUGAC ((((((((((.......))))))))))((((((((....(((.....)))......)))))))).....((((((((...(((((((((.......)))))....)))))))))))).. ( -27.70) >DroSim_CAF1 12876 119 - 1 CUUAUUGGAAAACGCAAUUUCAGUUAGAUCGAAUACAAAAACAUACCGUUAUCCUGUAUUCGAUCAAUCCAAUCGAUGUUUAGCUAUCGUACAAGCCGAUAACCCGCUAAUCGAUUGAC ...(((((((.......)))))))..(((((((((((..(((.....)))....)))))))))))....((((((((...(((((((((.......)))))....)))))))))))).. ( -31.40) >DroYak_CAF1 11736 104 - 1 CCAAUUGGAAAACUCUAUUUGAGUAAGAUCGAAUACAAAACCUUACCGUUAUUGUGCACUCGAUCAAUCUAAUCGAUUUUUAAAUAUCGAACAAGCC--UUA-------------UGAC ....(((((..((((.....))))..((((((.((((((((......))).)))))...))))))..)))))(((((........))))).......--...-------------.... ( -20.90) >consensus CUAAUUGGAAAACGCAAUUUCAGUUAGAUCGAAUACAAAAACAUACCGUUAUCCUGUAUUCGAUCAAUCCAAUCGAUGUUUAGCUAUCGAACAAGCCGAUAACCCGCUAAUCGAUUGAC .(((((((((.......)))))))))(((((((((((..(((.....)))....)))))))))))....((((((((.......(((((.......)))))........)))))))).. (-14.56 = -18.69 + 4.12)

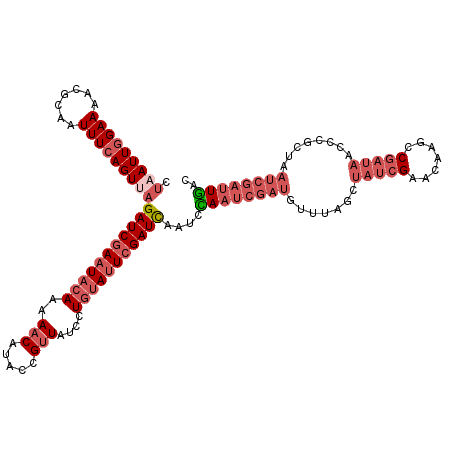

| Location | 5,609,524 – 5,609,632 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -19.07 |
| Energy contribution | -18.01 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609524 108 + 22224390 AAAUCGAUUAAAUUGAUCGAAUACAGGAUAACGGUACGUUUUUGUAUUAGAUCUAACUGAAACUACGUUUUCCAAUUAGA---------UUUCGUAUUAAUUGA-UUCUUGUUAAAAA- .((((((((((...((((.(((((((((..(((...)))))))))))).))))..........((((...((......))---------...))))))))))))-))...........- ( -21.10) >DroSec_CAF1 11198 108 + 1 ACAUCGAUUGGAUUAAUCGAAUAAAGGAUAACGGUAUGUUUUUGUAUUCGAUCUAACUGAAAUUGCGUGUUCCAAUUAGA---------UUUCGCAUUAAUCGA-UUCGUGUUAAGAA- (((.(((.(.((((((((((((((((((((......))))))).)))))))............((((...((......))---------...)))))))))).)-.))))))......- ( -22.80) >DroSim_CAF1 12916 108 + 1 ACAUCGAUUGGAUUGAUCGAAUACAGGAUAACGGUAUGUUUUUGUAUUCGAUCUAACUGAAAUUGCGUUUUCCAAUAAGA---------UUUCGCAUUAAUCGA-UUCGUGUUAAGAA- ..(((((((((...((((((((((((((...........))))))))))))))..........((((...((......))---------...))))))))))))-)............- ( -29.00) >DroYak_CAF1 11761 119 + 1 AAAUCGAUUAGAUUGAUCGAGUGCACAAUAACGGUAAGGUUUUGUAUUCGAUCUUACUCAAAUAGAGUUUUCCAAUUGGAACAUUGGAACCUUUCAGUAAUUGGAAGACUGAGAGCAUC .((((.....))))(((((((((((....(((......))).)))))))))))..((((.....)))).(((((((......))))))).((((((((.........)))))))).... ( -32.00) >consensus AAAUCGAUUAGAUUGAUCGAAUACAGGAUAACGGUAUGUUUUUGUAUUCGAUCUAACUGAAAUUGCGUUUUCCAAUUAGA_________UUUCGCAUUAAUCGA_UUCGUGUUAAGAA_ ...((((((((...((((((((((((((...........))))))))))))))...........(((.........................))).))))))))............... (-19.07 = -18.01 + -1.06)

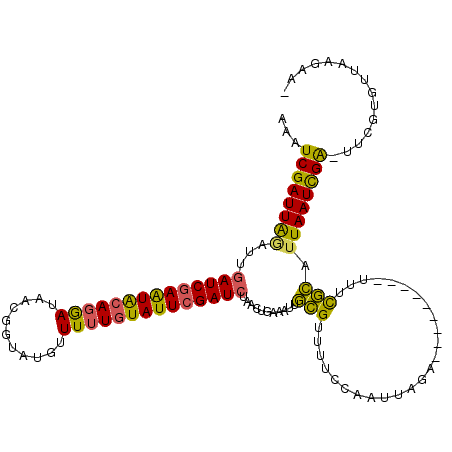
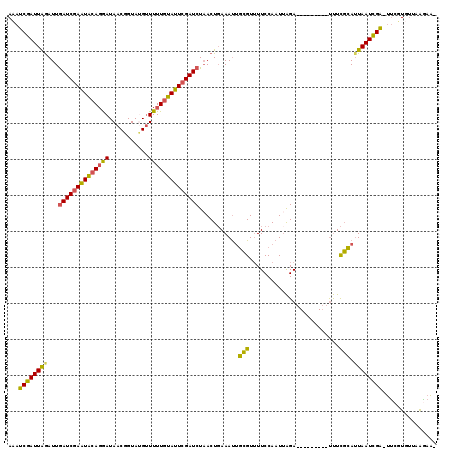
| Location | 5,609,524 – 5,609,632 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -11.84 |
| Energy contribution | -14.65 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609524 108 - 22224390 -UUUUUAACAAGAA-UCAAUUAAUACGAAA---------UCUAAUUGGAAAACGUAGUUUCAGUUAGAUCUAAUACAAAAACGUACCGUUAUCCUGUAUUCGAUCAAUUUAAUCGAUUU -..........(((-((.(((((...((..---------(((((((((((.......)))))))))))((.((((((..((((...))))....)))))).))))...))))).))))) ( -19.00) >DroSec_CAF1 11198 108 - 1 -UUCUUAACACGAA-UCGAUUAAUGCGAAA---------UCUAAUUGGAACACGCAAUUUCAGUUAGAUCGAAUACAAAAACAUACCGUUAUCCUUUAUUCGAUUAAUCCAAUCGAUGU -......((((((.-..(((((((.(((((---------(((((((((((.......))))))))))))..........(((.....)))........)))))))))))...))).))) ( -21.80) >DroSim_CAF1 12916 108 - 1 -UUCUUAACACGAA-UCGAUUAAUGCGAAA---------UCUUAUUGGAAAACGCAAUUUCAGUUAGAUCGAAUACAAAAACAUACCGUUAUCCUGUAUUCGAUCAAUCCAAUCGAUGU -............(-((((((..((((...---------(((....)))...))))..........(((((((((((..(((.....)))....))))))))))).....))))))).. ( -27.00) >DroYak_CAF1 11761 119 - 1 GAUGCUCUCAGUCUUCCAAUUACUGAAAGGUUCCAAUGUUCCAAUUGGAAAACUCUAUUUGAGUAAGAUCGAAUACAAAACCUUACCGUUAUUGUGCACUCGAUCAAUCUAAUCGAUUU (((.((.(((((.........))))).)).(((((((......))))))).((((.....))))..((((((.((((((((......))).)))))...))))))......)))..... ( -25.80) >consensus _UUCUUAACACGAA_UCAAUUAAUGCGAAA_________UCUAAUUGGAAAACGCAAUUUCAGUUAGAUCGAAUACAAAAACAUACCGUUAUCCUGUAUUCGAUCAAUCCAAUCGAUGU ...............((((((....................(((((((((.......)))))))))(((((((((((..(((.....)))....))))))))))).....))))))... (-11.84 = -14.65 + 2.81)

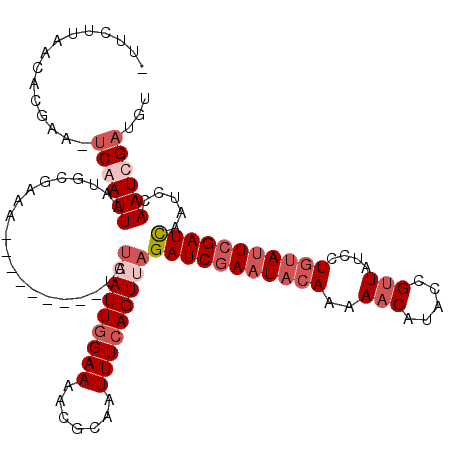
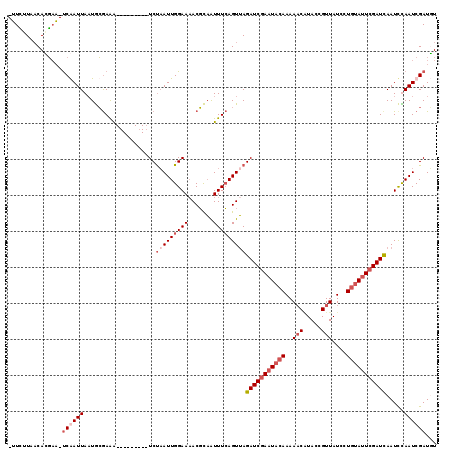
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:55 2006