
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,609,216 – 5,609,334 |
| Length | 118 |
| Max. P | 0.905455 |

| Location | 5,609,216 – 5,609,334 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -19.93 |
| Energy contribution | -20.37 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609216 118 + 22224390 GUGGUCCAGUUUUUUUUUUUGGCCACGCGCAGCGAUU-UUAUCUUAUCCGCUUUUCAUUCUCGCC-CGGAUAUAUAUUCCGCAUUAUUACCUGCUCUUCAGCUGAUCGUAUUUACCUUUC ((((.((((.........))))))))(((((((((..-.(((..((((((...............-)))))).))).)).(((........)))......))))..)))........... ( -23.46) >DroSec_CAF1 10901 108 + 1 GUGGCCCAGUUUUU--UUCUGGCCACGCGCAGCGAUU-UUAUCUUAUCCGCUUGUCUUUCUCGCC-CGGAUAUAUAUCCCGCAUUAUCACCUG--------CUGAUCGUAUUUACCUUUC ((((((.((.....--..)))))))).....((((((-......((((((..((.......))..-))))))........(((........))--------).))))))........... ( -25.30) >DroSim_CAF1 12619 108 + 1 GUGGCCCAGUUUUU--UUCUGGCCACGCGCAGCGAUU-UUAUCUUAUCCGCUUGUCUUUCUCGCC-CGGAUAUAUAUCCCGCAUUAUCACCUG--------CUGAUCGUAUUUACCUUUC ((((((.((.....--..)))))))).....((((((-......((((((..((.......))..-))))))........(((........))--------).))))))........... ( -25.30) >DroEre_CAF1 11337 118 + 1 GUGGCCCAGAUUUU--UCCUGGCCACGCGCAGCGAUUUUUAUCUUAUCCGCUCUUGUUUCACGCCCGGGAUAUAUAUCCAGCAUUAUCACCUGCCGUUCAGCUGAUCGUAUUUACCUUUC ((((((..((....--))..))))))((((((((((....)))...................(..((((((....)))).(((........)))))..).))))..)))........... ( -26.40) >DroYak_CAF1 11474 103 + 1 GUGGCCCAGAUUUU--UCCUGGCCACGCGCAGCGAUU-UUAUCUUAUCCGCC--------------CGGAUAUAUAUCCCGCAUUAUCACCUGCUGUUCAGCUGAUCGUAUUUACCUUUC ((((((..((....--))..))))))(((((((....-......(((((...--------------.)))))........(((........)))......))))..)))........... ( -26.70) >consensus GUGGCCCAGUUUUU__UUCUGGCCACGCGCAGCGAUU_UUAUCUUAUCCGCUUGUCUUUCUCGCC_CGGAUAUAUAUCCCGCAUUAUCACCUGC__UUCAGCUGAUCGUAUUUACCUUUC ((((.((((.........))))))))((((((((((....))).((((((................))))))............................))))..)))........... (-19.93 = -20.37 + 0.44)



| Location | 5,609,216 – 5,609,334 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.93 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -20.76 |
| Energy contribution | -22.56 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5609216 118 - 22224390 GAAAGGUAAAUACGAUCAGCUGAAGAGCAGGUAAUAAUGCGGAAUAUAUAUCCG-GGCGAGAAUGAAAAGCGGAUAAGAUAA-AAUCGCUGCGCGUGGCCAAAAAAAAAAACUGGACCAC ....(((...((((..((((.((...(((........)))........((((((-...............))))))......-..))))))..)))).(((...........)))))).. ( -22.36) >DroSec_CAF1 10901 108 - 1 GAAAGGUAAAUACGAUCAG--------CAGGUGAUAAUGCGGGAUAUAUAUCCG-GGCGAGAAAGACAAGCGGAUAAGAUAA-AAUCGCUGCGCGUGGCCAGAA--AAAAACUGGGCCAC ....(((...((((....(--------(((.((((....(.((((....)))).-)((...........))...........-.)))))))).)))).((((..--.....))))))).. ( -25.40) >DroSim_CAF1 12619 108 - 1 GAAAGGUAAAUACGAUCAG--------CAGGUGAUAAUGCGGGAUAUAUAUCCG-GGCGAGAAAGACAAGCGGAUAAGAUAA-AAUCGCUGCGCGUGGCCAGAA--AAAAACUGGGCCAC ....(((...((((....(--------(((.((((....(.((((....)))).-)((...........))...........-.)))))))).)))).((((..--.....))))))).. ( -25.40) >DroEre_CAF1 11337 118 - 1 GAAAGGUAAAUACGAUCAGCUGAACGGCAGGUGAUAAUGCUGGAUAUAUAUCCCGGGCGUGAAACAAGAGCGGAUAAGAUAAAAAUCGCUGCGCGUGGCCAGGA--AAAAUCUGGGCCAC ..............((((.(((.....))).)))).(((((((((....))))..))))).......(.((((....(((....))).)))).)((((((.(((--....))).)))))) ( -33.20) >DroYak_CAF1 11474 103 - 1 GAAAGGUAAAUACGAUCAGCUGAACAGCAGGUGAUAAUGCGGGAUAUAUAUCCG--------------GGCGGAUAAGAUAA-AAUCGCUGCGCGUGGCCAGGA--AAAAUCUGGGCCAC ..............((((.(((.....))).))))...(((((((....)))).--------------(((((.........-..))))).)))((((((.(((--....))).)))))) ( -30.80) >consensus GAAAGGUAAAUACGAUCAGCUGAA__GCAGGUGAUAAUGCGGGAUAUAUAUCCG_GGCGAGAAAGAAAAGCGGAUAAGAUAA_AAUCGCUGCGCGUGGCCAGAA__AAAAACUGGGCCAC ..............((((.(((.....))).))))...((.((((....))))................((((....(((....))).))))))((((((((.........)))).)))) (-20.76 = -22.56 + 1.80)

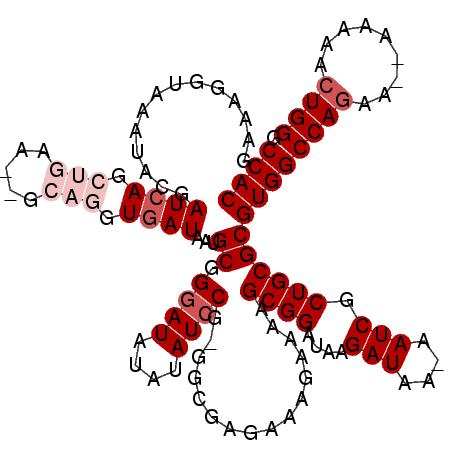

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:51 2006