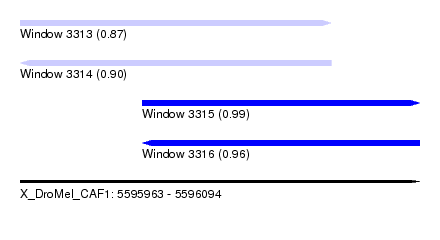
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,595,963 – 5,596,094 |
| Length | 131 |
| Max. P | 0.991932 |

| Location | 5,595,963 – 5,596,065 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -18.06 |
| Energy contribution | -16.58 |
| Covariance contribution | -1.48 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5595963 102 + 22224390 AAAGUGAAAUGCUGUAAGCCAUUUCCAGUGGCCUUUUGUCAUCACUGCCAGGGUAAAUAAACUUUCGUUUGCCUUUCUCCUGGUAUUGACACUUUGAAAUAU ((((((...........(((((.....))))).........(((.(((((((..((((((((....)))))..)))..))))))).)))))))))....... ( -27.70) >DroSec_CAF1 2834 102 + 1 AAAGUGAAAUGCUGUAAGCCAUUUCCUGUGACGUUUUGUCAUUUCUGCCAGGGUAAAUAAACUUUUGUUUACCCCUUCCCUGGUAUUGACUCUUUGAGAUAU ..((.(((((((.....).))))))))....((....((((....((((((((....(((((....)))))......)))))))).))))....))...... ( -25.10) >DroSim_CAF1 4642 102 + 1 AAAGUAAAAUGCUGUAAGACAUUUCCUGUGACCUUUUGUCAUCACUGCCAGGGUAAAUAAACUUACGUUUACCUCUUCCCUGGUAUUGACUCUUUGAAAUAU ....................(((((...((((.....))))(((.((((((((....(((((....)))))......)))))))).)))......))))).. ( -22.50) >DroEre_CAF1 2877 102 + 1 AAAGUGAAAAGUUAUAAGCCAUUCCCAUUGGCCUUUUGUCAUCACUGCAGGGGUAAACAAACUCUUGUUUGCCAUUCCACUCGUGUUGACUCUUUGAAAUAU .......((((......((((.......)))).....((((.(((....(((((((((((....)))))))))...))....))).)))).))))....... ( -23.20) >DroYak_CAF1 4857 102 + 1 UAAGUGAAAAGUUUUAAGCCAUUUUCAGUGGUCAUUUGCCAUCACUGCAAGGGUAAAUAAACUUUUGUUUGUCUUUCCUCUUGUAUUGACUCUUUGAAAUAU ..........((((((((((((.....))))).........(((.((((((((...((((((....))))))....)).)))))).)))....))))))).. ( -21.60) >consensus AAAGUGAAAUGCUGUAAGCCAUUUCCAGUGGCCUUUUGUCAUCACUGCCAGGGUAAAUAAACUUUUGUUUGCCUUUCCCCUGGUAUUGACUCUUUGAAAUAU ....................(((((...((((.....))))(((.((((((((....(((((....)))))......)))))))).)))......))))).. (-18.06 = -16.58 + -1.48)

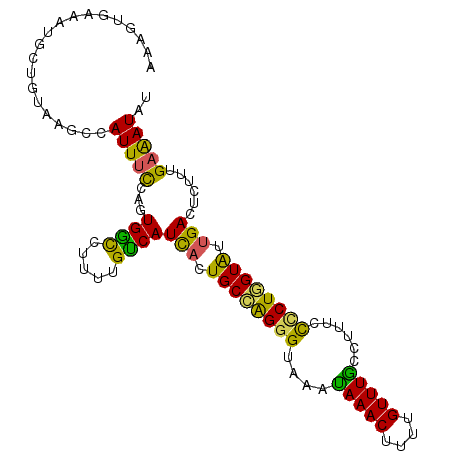
| Location | 5,595,963 – 5,596,065 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.73 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -17.46 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5595963 102 - 22224390 AUAUUUCAAAGUGUCAAUACCAGGAGAAAGGCAAACGAAAGUUUAUUUACCCUGGCAGUGAUGACAAAAGGCCACUGGAAAUGGCUUACAGCAUUUCACUUU .......(((((((((...(((((..(((...((((....)))).)))..)))))...))).......((((((.(....))))))).........)))))) ( -26.30) >DroSec_CAF1 2834 102 - 1 AUAUCUCAAAGAGUCAAUACCAGGGAAGGGGUAAACAAAAGUUUAUUUACCCUGGCAGAAAUGACAAAACGUCACAGGAAAUGGCUUACAGCAUUUCACUUU ..........((((((...((((((...((((((((....)))))))).))))))......((((.....)))).......))))))............... ( -25.20) >DroSim_CAF1 4642 102 - 1 AUAUUUCAAAGAGUCAAUACCAGGGAAGAGGUAAACGUAAGUUUAUUUACCCUGGCAGUGAUGACAAAAGGUCACAGGAAAUGUCUUACAGCAUUUUACUUU .......((((.((((...((((((...((((((((....)))))))).))))))...))))(((.....)))....(((((((......))))))).)))) ( -29.60) >DroEre_CAF1 2877 102 - 1 AUAUUUCAAAGAGUCAACACGAGUGGAAUGGCAAACAAGAGUUUGUUUACCCCUGCAGUGAUGACAAAAGGCCAAUGGGAAUGGCUUAUAACUUUUCACUUU .......((((.((((.(((.((.((...(((((((....)))))))...))))...))).))))...((((((.......))))))....))))....... ( -24.60) >DroYak_CAF1 4857 102 - 1 AUAUUUCAAAGAGUCAAUACAAGAGGAAAGACAAACAAAAGUUUAUUUACCCUUGCAGUGAUGGCAAAUGACCACUGAAAAUGGCUUAAAACUUUUCACUUA ..........((((((...((((.(((((...((((....)))).))).))))))(((((............)))))....))))))............... ( -16.70) >consensus AUAUUUCAAAGAGUCAAUACCAGGGGAAAGGCAAACAAAAGUUUAUUUACCCUGGCAGUGAUGACAAAAGGCCACUGGAAAUGGCUUACAGCAUUUCACUUU .......((((.((((.((((((((...((((((((....)))))))).))))))...)).))))............((((((........)))))).)))) (-17.46 = -18.14 + 0.68)



| Location | 5,596,003 – 5,596,094 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -17.96 |
| Energy contribution | -17.16 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5596003 91 + 22224390 AUCACUGCCAGGGUAAAUAAACUUUCGUUUGCCUUUCUCCUGGUAUUGACACUUUGAAAUAUUUAUUAUUUUUCUUUGUAGCAAAGUGGAG .(((.(((((((..((((((((....)))))..)))..))))))).)))(((((((..(((...............)))..)))))))... ( -24.06) >DroSec_CAF1 2874 91 + 1 AUUUCUGCCAGGGUAAAUAAACUUUUGUUUACCCCUUCCCUGGUAUUGACUCUUUGAGAUAUUUAUUAUUUCUCUUUGUAGCAAAGUGGAG .....((((((((....(((((....)))))......))))))))....((((..((((((.....)))))).(((((...))))).)))) ( -22.70) >DroSim_CAF1 4682 91 + 1 AUCACUGCCAGGGUAAAUAAACUUACGUUUACCUCUUCCCUGGUAUUGACUCUUUGAAAUAUUUAUUAUUUCUCUUUGCAGCAAAGUGGAG .(((.((((((((....(((((....)))))......)))))))).)))......((((((.....)))))).(((..(......)..))) ( -25.10) >DroEre_CAF1 2917 91 + 1 AUCACUGCAGGGGUAAACAAACUCUUGUUUGCCAUUCCACUCGUGUUGACUCUUUGAAAUAUUUAUUAUUUUUCUUCGUAGCAAAGUGGAG ...........(((((((((....)))))))))..((((((..(((((.......((((((.....))))))......))))).)))))). ( -24.22) >DroYak_CAF1 4897 91 + 1 AUCACUGCAAGGGUAAAUAAACUUUUGUUUGUCUUUCCUCUUGUAUUGACUCUUUGAAAUAUUUAUUAUUUUUCUUUGUAGCAAAGUGGAG .(((.((((((((...((((((....))))))....)).)))))).)))((((..((((((.....)))))).(((((...))))).)))) ( -18.70) >consensus AUCACUGCCAGGGUAAAUAAACUUUUGUUUGCCUUUCCCCUGGUAUUGACUCUUUGAAAUAUUUAUUAUUUUUCUUUGUAGCAAAGUGGAG .(((.((((((((....(((((....)))))......)))))))).)))((((..((((((.....)))))).(((((...))))).)))) (-17.96 = -17.16 + -0.80)



| Location | 5,596,003 – 5,596,094 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -19.06 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5596003 91 - 22224390 CUCCACUUUGCUACAAAGAAAAAUAAUAAAUAUUUCAAAGUGUCAAUACCAGGAGAAAGGCAAACGAAAGUUUAUUUACCCUGGCAGUGAU ...(((((((..........(((((.....))))))))))))(((...(((((..(((...((((....)))).)))..)))))...))). ( -20.30) >DroSec_CAF1 2874 91 - 1 CUCCACUUUGCUACAAAGAGAAAUAAUAAAUAUCUCAAAGAGUCAAUACCAGGGAAGGGGUAAACAAAAGUUUAUUUACCCUGGCAGAAAU ......(((((......((((.((.....)).)))).............(((((...((((((((....)))))))).))))))))))... ( -21.20) >DroSim_CAF1 4682 91 - 1 CUCCACUUUGCUGCAAAGAGAAAUAAUAAAUAUUUCAAAGAGUCAAUACCAGGGAAGAGGUAAACGUAAGUUUAUUUACCCUGGCAGUGAU .......(..((((.....((((((.....)))))).............(((((...((((((((....)))))))).)))))))))..). ( -28.60) >DroEre_CAF1 2917 91 - 1 CUCCACUUUGCUACGAAGAAAAAUAAUAAAUAUUUCAAAGAGUCAACACGAGUGGAAUGGCAAACAAGAGUUUGUUUACCCCUGCAGUGAU .(((((((((((.(...((((.((.....)).))))...))).))....)))))))..(((((((....)))))))............... ( -14.00) >DroYak_CAF1 4897 91 - 1 CUCCACUUUGCUACAAAGAAAAAUAAUAAAUAUUUCAAAGAGUCAAUACAAGAGGAAAGACAAACAAAAGUUUAUUUACCCUUGCAGUGAU .....(((((...))))).......................((((...((((.(((((...((((....)))).))).))))))...)))) ( -11.20) >consensus CUCCACUUUGCUACAAAGAAAAAUAAUAAAUAUUUCAAAGAGUCAAUACCAGGGGAAAGGCAAACAAAAGUUUAUUUACCCUGGCAGUGAU .....(((((...))))).......................((((...((((((...((((((((....)))))))).))))))...)))) (-14.96 = -15.64 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:45 2006