
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,583,698 – 5,583,857 |
| Length | 159 |
| Max. P | 0.771843 |

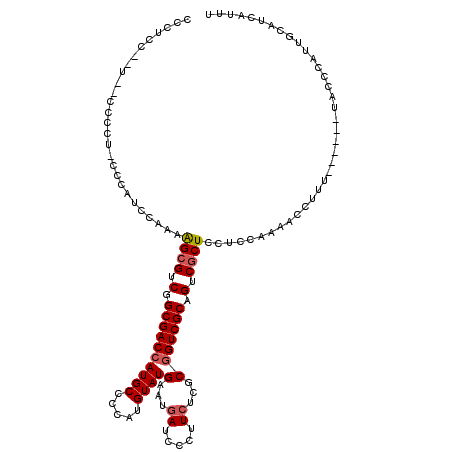
| Location | 5,583,698 – 5,583,817 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5583698 119 - 22224390 CCCUCCACUC-CCCCUGCCCAUACAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCAGUCGCUCCUCCAAAGCCUUUUACUUUUACCCAUUGCAUCAUUU ..........-....(((.........((((.(.((((((((((.....))))(...((.....))...))))))).).)))).....((((.......))))........)))...... ( -19.80) >DroSec_CAF1 11826 106 - 1 CCCUCC-----CCC---CUCAUCAAAAAGCGUCGGCGACCAUGCCACAUGUAUGAAUGAUCCCUUCUCGCGGUCGCCGUCGCUCCUCCAAAACCUUU------AACCCAUUGCAUCAUUU ......-----...---..........((((.((((((((((((.....))))(...((.....))...))))))))).))))..............------................. ( -25.00) >DroSim_CAF1 11951 106 - 1 CCCUCC-----CCC---CUCAUCAAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCCGUCACUCCUCCAAAACCUUU------AACCCAUUGCAUCAUUU ......-----...---..........((.(.((((((((((((.....))))(...((.....))...))))))))).).))..............------................. ( -19.60) >DroEre_CAF1 12285 111 - 1 UUCUUAUAUG-CCCCUCCCGAUCCAAAGGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUU--CGCAGUCGCAGUCGCUCCUCCAAACCCUUU------UGCCCAUUGUAUCAUUU ..........-........(((.(((.((((.(.((((((((((.....)))))...((.....)--)...))))).).))))....((((....))------))....))).))).... ( -20.40) >DroYak_CAF1 12344 114 - 1 CACCCACCUCCCCCCUCCCCAUCCAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCAGUCGCUGCUCCAAAACCAUU------UACCCAUUUUAUUUUUU ...........................((((.(.((((((((((.....))))(...((.....))...))))))).).))))..............------................. ( -19.50) >consensus CCCUCC__U__CCCCU_CCCAUCCAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCAGUCGCUCCUCCAAAACCUUU______UACCCAUUGCAUCAUUU ...........................((((.(.((((((((((.....))))(...((.....))...))))))).).))))..................................... (-15.46 = -15.90 + 0.44)

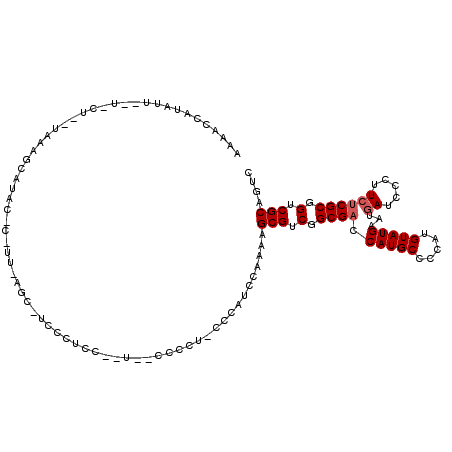
| Location | 5,583,738 – 5,583,857 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.11 |
| Mean single sequence MFE | -17.21 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5583738 119 - 22224390 AAAACCAUAUAUUUUCUCAUAAGGCAUACCCUUUCAGCUUCCCUCCACUC-CCCCUGCCCAUACAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCAGUC ....................((((.....)))).................-...((((.((((((...((((.(....).))))....))))))...((((.(.....).)))))))).. ( -18.70) >DroSec_CAF1 11860 104 - 1 AAAACCAUAUU--------UAAAGCAUACUCCUUUAGCACCCCUCC-----CCC---CUCAUCAAAAAGCGUCGGCGACCAUGCCACAUGUAUGAAUGAUCCCUUCUCGCGGUCGCCGUC ..........(--------(((((.......)))))).........-----...---.............(.((((((((((((.....))))(...((.....))...))))))))).) ( -18.30) >DroSim_CAF1 11985 104 - 1 AAAUCCAUAUU--------UAAAGCAUACUCCUUUAGCACCCCUCC-----CCC---CUCAUCAAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCCGUC ..........(--------(((((.......)))))).........-----...---.............(.((((((((((((.....))))(...((.....))...))))))))).) ( -18.30) >DroEre_CAF1 12319 95 - 1 -----UAUAUC--UCCUUUUAUCU---------------UUUCUUAUAUG-CCCCUCCCGAUCCAAAGGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUU--CGCAGUCGCAGUC -----......--...........---------------........(((-((..............)))))..((((((((((.....)))))...((.....)--)...))))).... ( -14.84) >DroYak_CAF1 12378 105 - 1 AUAAGCAUAUU--UUCUCAUAAGUCA-------------UCACCCACCUCCCCCCUCCCCAUCCAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCAGUC ...........--.............-------------.............................(((.(.((((.(((((.....)))))...((.....)))))).).))).... ( -15.90) >consensus AAAACCAUAUU__U_CU__UAAAGCAUAC_C_UU_AGC_UCCCUCC__U__CCCCU_CCCAUCCAAAAGCGUCGGCGACCAUGCCCCAUGUAUGAAUGAUCCCUUCUCGCGGUCGCAGUC ....................................................................(((.(.((((.(((((.....)))))...((.....)))))).).))).... (-13.32 = -13.72 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:36 2006