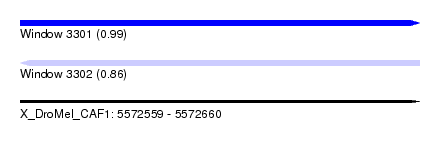
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,572,559 – 5,572,660 |
| Length | 101 |
| Max. P | 0.990609 |

| Location | 5,572,559 – 5,572,660 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.83 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -18.24 |
| Energy contribution | -17.76 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5572559 101 + 22224390 -GCCGACCUCC------------GUCUGAUGAAUCCAAGUUCAUUUCCAAAGUGCAUUACUUGGUUGCCCCAUUAAG---ACGGGCA--AGAAGUUGAACAACUUCCAAUUAUCAU-CAU -...(((....------------)))((((((..((((((.((((.....))))....))))))((((((.......---..)))))--)((((((....))))))......))))-)). ( -27.20) >DroSec_CAF1 5399 92 + 1 -GCCGAC-----------------------GCAUCCAAGUUCACUUCCCAAGUGCAUUACUUGGUUGCCCCUUGCAG---UUGGGCAGAAGAAGUUGAACAACUUCCAAUUAUCAU-CAU -((....-----------------------))......((((((((((((((((...)))))))((((((.......---..))))))..)))).)))))................-... ( -24.00) >DroSim_CAF1 5480 92 + 1 -GCCGAC-----------------------GCAUCCAAGUUCACUUCCCAAGUGCAUUACUUGGUUGCCCCUUUCAG---AUGGGCAGAAGAAGUUGAACAACUUCCAAUUAUCAU-CAU -......-----------------------(((.((((((.((((.....))))....)))))).)))........(---((((......((((((....))))))......))))-).. ( -24.80) >DroEre_CAF1 5740 110 + 1 -GGCCACCCACAGUCUACAUAGAGUCUGACGAAUCCAAGUUCACUUC-AAAGUGCAUUGCUUAGUUGGCCCAUUAAA---UCGGCCA--AGAAGUUC-ACAACUUCCA-UUAUUAU-CAU -((((.....(((.((......)).))).(((....((((.((((..-..))))....))))..)))..........---..)))).--.(((((..-...)))))..-.......-... ( -20.70) >DroYak_CAF1 5700 113 + 1 UCGACCUCCAGAGUCUAC----AGUCUAACGAAUCCAAGUUCACUUC-AAAGUGCAUUGCUUAGUUGGCCCAUUGAAUCAUCGGCCA--AGAAGUUGAACAACUUCCAAUUAUUAUCCAU ..((((....).)))...----..((....))......(((((((((-.((((.....))))..((((((.((......)).)))))--))))).))))).................... ( -22.30) >consensus _GCCGACC_______________GUCU_A_GAAUCCAAGUUCACUUCCAAAGUGCAUUACUUGGUUGCCCCAUUAAG___UCGGGCA__AGAAGUUGAACAACUUCCAAUUAUCAU_CAU ..................................((((((.((((.....))))....)))))).(((((............)))))...((((((....)))))).............. (-18.24 = -17.76 + -0.48)

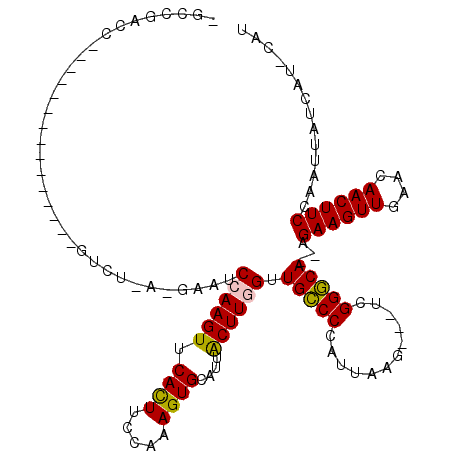
| Location | 5,572,559 – 5,572,660 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.83 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.88 |
| Energy contribution | -20.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5572559 101 - 22224390 AUG-AUGAUAAUUGGAAGUUGUUCAACUUCU--UGCCCGU---CUUAAUGGGGCAACCAAGUAAUGCACUUUGGAAAUGAACUUGGAUUCAUCAGAC------------GGAGGUCGGC- .((-((((.....(((((((....)))))))--(((((..---.......))))).((((((.((...........))..))))))..))))))(((------------....)))...- ( -27.90) >DroSec_CAF1 5399 92 - 1 AUG-AUGAUAAUUGGAAGUUGUUCAACUUCUUCUGCCCAA---CUGCAAGGGGCAACCAAGUAAUGCACUUGGGAAGUGAACUUGGAUGC-----------------------GUCGGC- .((-(((...(((.((....(((((.((((...(((((..---.......))))).((((((.....))))))))))))))))).))).)-----------------------))))..- ( -28.60) >DroSim_CAF1 5480 92 - 1 AUG-AUGAUAAUUGGAAGUUGUUCAACUUCUUCUGCCCAU---CUGAAAGGGGCAACCAAGUAAUGCACUUGGGAAGUGAACUUGGAUGC-----------------------GUCGGC- .((-(((...(((.((....(((((.((((...(((((..---.......))))).((((((.....))))))))))))))))).))).)-----------------------))))..- ( -28.60) >DroEre_CAF1 5740 110 - 1 AUG-AUAAUAA-UGGAAGUUGU-GAACUUCU--UGGCCGA---UUUAAUGGGCCAACUAAGCAAUGCACUUU-GAAGUGAACUUGGAUUCGUCAGACUCUAUGUAGACUGUGGGUGGCC- ...-.......-.((((((...-..))))))--(((((.(---(...)).)))))...........(((((.-..(((..((.((((.((....)).)))).))..)))..)))))...- ( -25.30) >DroYak_CAF1 5700 113 - 1 AUGGAUAAUAAUUGGAAGUUGUUCAACUUCU--UGGCCGAUGAUUCAAUGGGCCAACUAAGCAAUGCACUUU-GAAGUGAACUUGGAUUCGUUAGACU----GUAGACUCUGGAGGUCGA ((((((.......(((((((....)))))))--(((((.((......)).))))).(((((.....((((..-..))))..)))))))))))..((((----.(((...)))..)))).. ( -28.50) >consensus AUG_AUGAUAAUUGGAAGUUGUUCAACUUCU__UGCCCGA___CUGAAUGGGGCAACCAAGUAAUGCACUUUGGAAGUGAACUUGGAUUC_U_AGAC_______________GGUCGGC_ .............(((((((....)))))))..(((((............))))).((((((....((((.....)))).)))))).................................. (-20.88 = -20.76 + -0.12)

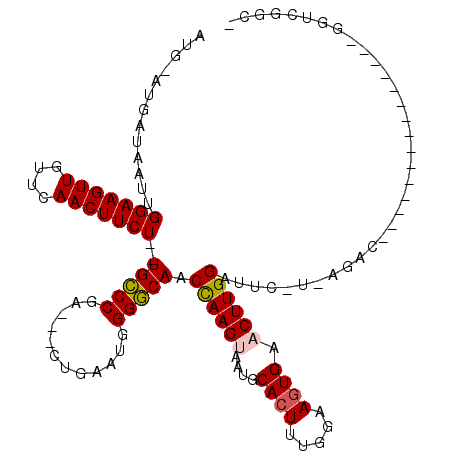
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:33 2006