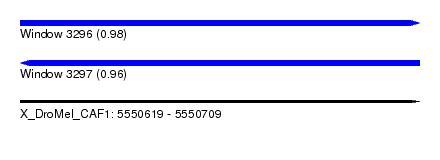
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,550,619 – 5,550,709 |
| Length | 90 |
| Max. P | 0.980141 |

| Location | 5,550,619 – 5,550,709 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.78 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5550619 90 + 22224390 --------UAUGACUAAGAACUUGUUGGUUAAUAAUCGUAGCAUAGUUUUUGGCGCUAUGUUGGGGUUUGUUGUACUAAGUUGGAGCCACAAAACUUU --------(((((.((..((((....))))..)).)))))....((((((((((.(((..((((...........))))..))).)))).)))))).. ( -18.80) >DroSec_CAF1 6097 90 + 1 --------UAUGACUAAGAACUAGUUGGUUAAUAAUCAUAGCAUAGUUUUUGGCGCUAAGUUGGGGUUUAUAGUACAAACUUAGUACCACAGAACUUU --------(((((.((..((((....))))..)).)))))....(((((((((..(((((((...((.......)).)))))))..))).)))))).. ( -21.90) >DroYak_CAF1 8658 84 + 1 AUGUAAAAUAUGGCUAAGAACUCGUUGGCUAAAAAUCAUAGCAUAGUUUUUGGCGCUAAGUUGGGC--------------UUAGUGCCUAAGAACUUU ..........((((((((....).))))))).............((((((((((((((((.....)--------------)))))))).))))))).. ( -25.60) >consensus ________UAUGACUAAGAACUAGUUGGUUAAUAAUCAUAGCAUAGUUUUUGGCGCUAAGUUGGGGUUU_U_GUAC_AA_UUAGUGCCACAGAACUUU ..........(((((((.......))))))).............(((((((((((((((.....................))))))))).)))))).. (-13.38 = -13.83 + 0.45)


| Location | 5,550,619 – 5,550,709 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Mean single sequence MFE | -16.49 |
| Consensus MFE | -8.87 |
| Energy contribution | -10.43 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5550619 90 - 22224390 AAAGUUUUGUGGCUCCAACUUAGUACAACAAACCCCAACAUAGCGCCAAAAACUAUGCUACGAUUAUUAACCAACAAGUUCUUAGUCAUA-------- ..((((((.((((...............................)))))))))).......(((((..(((......)))..)))))...-------- ( -12.08) >DroSec_CAF1 6097 90 - 1 AAAGUUCUGUGGUACUAAGUUUGUACUAUAAACCCCAACUUAGCGCCAAAAACUAUGCUAUGAUUAUUAACCAACUAGUUCUUAGUCAUA-------- ..((((...(((..(((((((.((.......))...)))))))..)))..))))....((((((((..(((......)))..))))))))-------- ( -20.40) >DroYak_CAF1 8658 84 - 1 AAAGUUCUUAGGCACUAA--------------GCCCAACUUAGCGCCAAAAACUAUGCUAUGAUUUUUAGCCAACGAGUUCUUAGCCAUAUUUUACAU .(((..(((.(((.((((--------------(.....))))).))).........((((.......))))....)))..)))............... ( -17.00) >consensus AAAGUUCUGUGGCACUAA_UU_GUAC_A_AAACCCCAACUUAGCGCCAAAAACUAUGCUAUGAUUAUUAACCAACAAGUUCUUAGUCAUA________ ..((((...((((.((((.....................)))).))))..))))....((((((((..(((......)))..))))))))........ ( -8.87 = -10.43 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:29 2006