
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,543,118 – 5,543,434 |
| Length | 316 |
| Max. P | 0.989692 |

| Location | 5,543,118 – 5,543,234 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -18.77 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5543118 116 + 22224390 CUUCACUUUGAAGAGUAAAAUUUUGAUUUCGAACAUGAUGUUACCAAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAA----UUUCUAGUUGAAAUAUAA ((((.....))))............(((((((....((.(((.........((((......))))....(((((((((.....)))))))))....))----).))...))))))).... ( -25.20) >DroSec_CAF1 5031 116 + 1 CUCCA----AAAGAGUCCAAUUUUGAUUUCGAAAACGAUGUUACCGAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAAUAAAUAUCAAGUAGAAAAAUAA ...((----(((........))))).((((......((((((.........((((......))))....(((((((((.....)))))))))........)))))).....))))..... ( -23.70) >DroSim_CAF1 4788 116 + 1 CUUCACUUUGAAGAGUCCAAUUAUGGUUUCGAAAACGAUGUUACCGAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAA----UUUCAAGUAGAAAAGUAA .(((((((.((((..........((((((((....))).(..((((....))))..)....)))))...(((((((((.....)))))))))......----)))))))).)))...... ( -27.60) >consensus CUUCACUUUGAAGAGUCCAAUUUUGAUUUCGAAAACGAUGUUACCGAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAA____UUUCAAGUAGAAAAAUAA ((((.....)))).............((((.....................((((......))))....(((((((((.....)))))))))...................))))..... (-18.77 = -19.43 + 0.67)

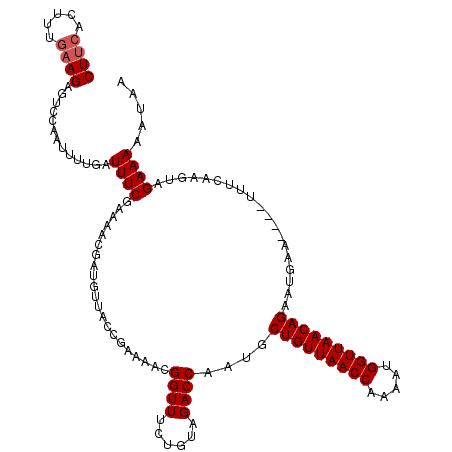
| Location | 5,543,118 – 5,543,234 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -19.81 |
| Energy contribution | -21.03 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5543118 116 - 22224390 UUAUAUUUCAACUAGAAA----UUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUUGGUAACAUCAUGUUCGAAAUCAAAAUUUUACUCUUCAAAGUGAAG ....(((((.((((((((----(...((((((..((((..(((.....)))...))))..))))))...)))))))))(((.....))).)))))......((((((......)))))). ( -24.20) >DroSec_CAF1 5031 116 - 1 UUAUUUUUCUACUUGAUAUUUAUUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUCGGUAACAUCGUUUUCGAAAUCAAAAUUGGACUCUUU----UGGAG .......((((...((........((..(((((((((.....)))))))))...))(((((.....((((....))))....(((....))).........)))))))...----)))). ( -24.50) >DroSim_CAF1 4788 116 - 1 UUACUUUUCUACUUGAAA----UUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUCGGUAACAUCGUUUUCGAAACCAUAAUUGGACUCUUCAAAGUGAAG ......(((.(((((((.----......(((((((((.....)))))))))....((((((.....((((....))))....(((....))).........)))))).))).))))))). ( -27.20) >consensus UUAUUUUUCUACUUGAAA____UUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUCGGUAACAUCGUUUUCGAAAUCAAAAUUGGACUCUUCAAAGUGAAG ......................(((((((((((((((.....)))))))))....((((((.....((((....))))....(((....))).........))))))......)))))). (-19.81 = -21.03 + 1.23)


| Location | 5,543,158 – 5,543,274 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -21.69 |
| Energy contribution | -21.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5543158 116 + 22224390 UUACCAAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAA----UUUCUAGUUGAAAUAUAAAUAAAAGGAAAGCAAAGAAAGCCUCCAGCUAAUGGGUUUU ......((((((((((((.....((....(((((((((.....))))))))).....(----((((.....)))))..........)).......)).))))).(((.....)))))))) ( -25.60) >DroSec_CAF1 5067 120 + 1 UUACCGAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAAUAAAUAUCAAGUAGAAAAAUAAACAAAAGGAAAGGAAAGAAAGCCUCCAGCAAAUGGGUCUU ..((((....))))......(((((..(((((((((((.....)))))))....................................(((..((........)))))))))....))))). ( -25.90) >DroSim_CAF1 4828 116 + 1 UUACCGAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAA----UUUCAAGUAGAAAAGUAAAUAAAGGGAAAGGAAAGAAAGCCUCCUGCAAAUGGGUCUU ..((((....))))......(((((....(((((((((.....)))))))))......----((((.....))))..........((((..((........)))))).......))))). ( -26.90) >consensus UUACCGAAAACGGUUUCUGUAGACCAAUGCUGUUAACCAAAAUGGUUAACAGAAUGAA____UUUCAAGUAGAAAAAUAAAUAAAAGGAAAGGAAAGAAAGCCUCCAGCAAAUGGGUCUU ..(((......)))......(((((....(((((((((.....)))))))))..................................(((..((........)))))........))))). (-21.69 = -21.80 + 0.11)

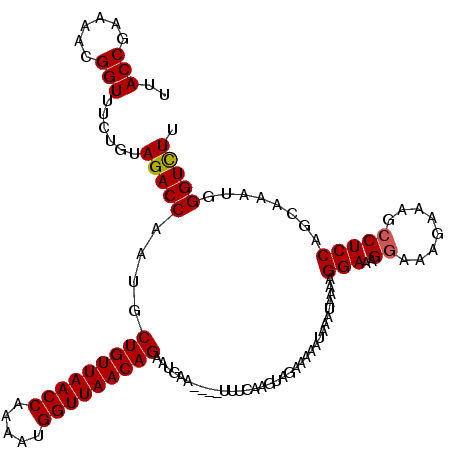
| Location | 5,543,158 – 5,543,274 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5543158 116 - 22224390 AAAACCCAUUAGCUGGAGGCUUUCUUUGCUUUCCUUUUAUUUAUAUUUCAACUAGAAA----UUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUUGGUAA .....(((..(((.((((((.......))))))............((((...((((..----.(((..(((((((((.....)))))))))...)))))))..))))..)))..)))... ( -26.90) >DroSec_CAF1 5067 120 - 1 AAGACCCAUUUGCUGGAGGCUUUCUUUCCUUUCCUUUUGUUUAUUUUUCUACUUGAUAUUUAUUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUCGGUAA .(((((....(((((((((.....))))).........................................(((((((.....)))))))))))..)))))......((((....)))).. ( -26.60) >DroSim_CAF1 4828 116 - 1 AAGACCCAUUUGCAGGAGGCUUUCUUUCCUUUCCCUUUAUUUACUUUUCUACUUGAAA----UUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUCGGUAA .(((((....(((((((((.....))))))...............((((.....))))----.......((((((((.....)))))))))))..)))))......((((....)))).. ( -28.00) >consensus AAGACCCAUUUGCUGGAGGCUUUCUUUCCUUUCCUUUUAUUUAUUUUUCUACUUGAAA____UUCAUUCUGUUAACCAUUUUGGUUAACAGCAUUGGUCUACAGAAACCGUUUUCGGUAA .(((((........(((((..........)))))..................................(((((((((.....)))))))))....)))))......((((....)))).. (-22.25 = -22.37 + 0.11)


| Location | 5,543,274 – 5,543,394 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -42.97 |
| Consensus MFE | -40.99 |
| Energy contribution | -41.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5543274 120 + 22224390 GGGCUAUCCAAACACUUUUUGGCCAGCUGAUUGGCCAAGGACACAUGCGCUUUAUUUUCACUGUUAACGCGAAGAGCGCUCUGUAUUGGCCAAGCCAACUUCGACAUCGAACCGGGCUCG .((((..(((((.....)))))..))))..((((((((..(((...((((((....(((.(.......).)))))))))..))).))))))))(((...((((....))))...)))... ( -40.70) >DroSec_CAF1 5187 120 + 1 GGGCUAUCCAAACUCUUUUUGGCCAGCUGAUUGGCCAAGGACACAUGCGCUUUAUUUUCACUGUUAACGCGAAGAGCGCUCUGUCUUGGCCAAGCCAAGUUCGACAUCGAACCGGGCUCG .((((..(((((.....)))))..))))..(((((((((.(((...((((((....(((.(.......).)))))))))..))))))))))))(((..(((((....)))))..)))... ( -46.90) >DroSim_CAF1 4944 120 + 1 GAUCUAUCCAAACUCUUUUUGGCCAGCUGAUUGGCCAAGGACACAUGCGCUUUAAUUUCACUGUUAACGCGAAGAGCGCUCUGUCUUGGCCAAGCCAAGUUCGACAUCGAACCGGGCUCG ...((..(((((.....)))))..))....(((((((((.(((...((((((....(((.(.......).)))))))))..))))))))))))(((..(((((....)))))..)))... ( -41.30) >consensus GGGCUAUCCAAACUCUUUUUGGCCAGCUGAUUGGCCAAGGACACAUGCGCUUUAUUUUCACUGUUAACGCGAAGAGCGCUCUGUCUUGGCCAAGCCAAGUUCGACAUCGAACCGGGCUCG .((((..(((((.....)))))..))))..(((((((((.(((...((((((....(((.(.......).)))))))))..))))))))))))(((..(((((....)))))..)))... (-40.99 = -41.77 + 0.78)

| Location | 5,543,274 – 5,543,394 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -48.47 |
| Consensus MFE | -46.53 |
| Energy contribution | -47.53 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5543274 120 - 22224390 CGAGCCCGGUUCGAUGUCGAAGUUGGCUUGGCCAAUACAGAGCGCUCUUCGCGUUAACAGUGAAAAUAAAGCGCAUGUGUCCUUGGCCAAUCAGCUGGCCAAAAAGUGUUUGGAUAGCCC .(((((.(.((((....)))).).)))))((((((((((..(((((.(((((.......))))).....))))).)))))...))))).....((((.(((((.....))))).)))).. ( -44.30) >DroSec_CAF1 5187 120 - 1 CGAGCCCGGUUCGAUGUCGAACUUGGCUUGGCCAAGACAGAGCGCUCUUCGCGUUAACAGUGAAAAUAAAGCGCAUGUGUCCUUGGCCAAUCAGCUGGCCAAAAAGAGUUUGGAUAGCCC .(((((.((((((....)))))).)))))((((((((((.((((((.(((((.......))))).....))))).).))).))))))).....((((.(((((.....))))).)))).. ( -52.10) >DroSim_CAF1 4944 120 - 1 CGAGCCCGGUUCGAUGUCGAACUUGGCUUGGCCAAGACAGAGCGCUCUUCGCGUUAACAGUGAAAUUAAAGCGCAUGUGUCCUUGGCCAAUCAGCUGGCCAAAAAGAGUUUGGAUAGAUC .(((((.((((((....)))))).)))))((((((((((.((((((.(((((.......))))).....))))).).))).)))))))......(((.(((((.....))))).)))... ( -49.00) >consensus CGAGCCCGGUUCGAUGUCGAACUUGGCUUGGCCAAGACAGAGCGCUCUUCGCGUUAACAGUGAAAAUAAAGCGCAUGUGUCCUUGGCCAAUCAGCUGGCCAAAAAGAGUUUGGAUAGCCC .(((((.((((((....)))))).)))))((((((((((.((((((.(((((.......))))).....))))).).))).))))))).....((((.(((((.....))))).)))).. (-46.53 = -47.53 + 1.00)

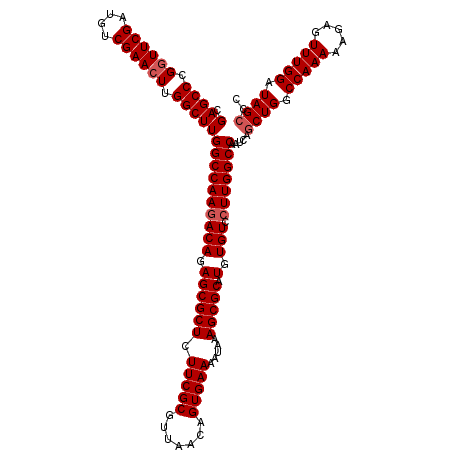
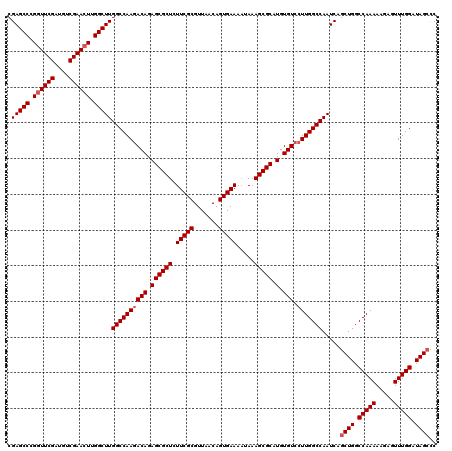
| Location | 5,543,314 – 5,543,434 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -41.87 |
| Consensus MFE | -39.80 |
| Energy contribution | -40.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5543314 120 - 22224390 AGCACUUGCUCAGCACUGGCUUGAUCUCUUGCAACCGGUUCGAGCCCGGUUCGAUGUCGAAGUUGGCUUGGCCAAUACAGAGCGCUCUUCGCGUUAACAGUGAAAAUAAAGCGCAUGUGU .((((.(((...((.(((((..(....)..))....(((.((((((.(.((((....)))).).)))))))))....))).))(((.(((((.......))))).....)))))).)))) ( -40.30) >DroSec_CAF1 5227 120 - 1 AGCACUUGCUCAGCACUGGCUUGAUCUCUCGCAACAGGUUCGAGCCCGGUUCGAUGUCGAACUUGGCUUGGCCAAGACAGAGCGCUCUUCGCGUUAACAGUGAAAAUAAAGCGCAUGUGU .((((.(((...((.(((((..(....)..))....(((.((((((.((((((....)))))).)))))))))....))).))(((.(((((.......))))).....)))))).)))) ( -43.20) >DroSim_CAF1 4984 120 - 1 AGCACUUGCUCAGCACUGACUUGAUCUCUUGCAACAGGUUCGAGCCCGGUUCGAUGUCGAACUUGGCUUGGCCAAGACAGAGCGCUCUUCGCGUUAACAGUGAAAUUAAAGCGCAUGUGU .((((.(((...((.(((.((((....((((...)))).(((((((.((((((....)))))).))))))).)))).))).))(((.(((((.......))))).....)))))).)))) ( -42.10) >consensus AGCACUUGCUCAGCACUGGCUUGAUCUCUUGCAACAGGUUCGAGCCCGGUUCGAUGUCGAACUUGGCUUGGCCAAGACAGAGCGCUCUUCGCGUUAACAGUGAAAAUAAAGCGCAUGUGU .((((.(((...((.(((((..(....)..))....(((.((((((.((((((....)))))).)))))))))....))).))(((.(((((.......))))).....)))))).)))) (-39.80 = -40.47 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:26 2006