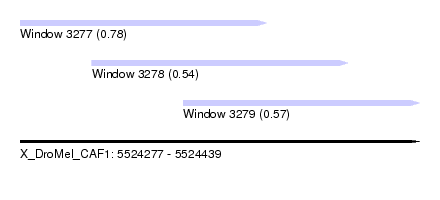
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,524,277 – 5,524,439 |
| Length | 162 |
| Max. P | 0.784235 |

| Location | 5,524,277 – 5,524,377 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -7.15 |
| Energy contribution | -9.46 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5524277 100 + 22224390 CUUUCAUUUCCAUUUCCAAUUUAUUUUUCCCAAGUCUGGCUCAUUUUGUGCAAAUAAACAAAUGACUCAUUUGUUUAAAAAUU-CGAUGAGACUCAAGGAA----- .............((((............(((....)))((((((..((.....((((((((((...))))))))))...)).-.))))))......))))----- ( -17.50) >DroSec_CAF1 14153 92 + 1 CUUUCAUUUCCAUU-UCAAUUUUUUUUUCCA-AGUCUGACUCAUUUUAGGCAAAUAAACAAAUGACUCAUUUGUUCCAAAUUU-AAAUGAGACUC----------- .((((((((..(((-(...............-.((((((......)))))).....((((((((...))))))))..))))..-))))))))...----------- ( -14.60) >DroSim_CAF1 16690 94 + 1 CUUUCAUUUCUAUUUUUAAUUUAUUUUUCCAAAGUCUGGCUCAUUUUAGGCAAAUAAACAAAUGACUCAUUUGUUCCAAAUUU-AAAUGAGACUC----------- .((((((((..((((..................((((((......)))))).....((((((((...))))))))..))))..-))))))))...----------- ( -14.70) >DroEre_CAF1 14359 99 + 1 CUUUCAUUUCAAUUGCAAAUGUAUUUUUCCA-GGACAUGAUCCUAUUAUGGAAAGAAACAAAUGACUCAUUUGUUUCAAAAUUUCGGC------CAAACAAUGAAU ..((((((......((.........((((((-(((.....))).....))))))((((((((((...)))))))))).........))------.....)))))). ( -19.90) >consensus CUUUCAUUUCCAUUUCCAAUUUAUUUUUCCA_AGUCUGGCUCAUUUUAGGCAAAUAAACAAAUGACUCAUUUGUUCCAAAAUU_AAAUGAGACUC___________ .......................................(((((((........((((((((((...)))))))))).......)))))))............... ( -7.15 = -9.46 + 2.31)


| Location | 5,524,306 – 5,524,410 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 63.58 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -5.19 |
| Energy contribution | -7.23 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5524306 104 + 22224390 CCAAGUCUGGCUCAUUUUGUGCAAAUAAACAAAUGACUCAUUUGUUUAAAAAUU-CGAUGAGACUCAAGGAA-----UGUC-ACAUUCACAAA-AAUUUUCAAUUUGAAAUC----- .(((((.(((....(((((((....((((((((((...))))))))))......-.((((.(((........-----.)))-.))))))))))-)....)))))))).....----- ( -19.90) >DroSec_CAF1 14181 93 + 1 CA-AGUCUGACUCAUUUUAGGCAAAUAAACAAAUGACUCAUUUGUUCCAAAUUU-AAAUGAGACUC----------------ACAUUCACAAA-UAUUUUCAAUUUGAAAUC----- ..-.((((((......))))))..............((((((((.........)-)))))))....----------------.......((((-(.......))))).....----- ( -13.10) >DroSim_CAF1 16719 93 + 1 CAAAGUCUGGCUCAUUUUAGGCAAAUAAACAAAUGACUCAUUUGUUCCAAAUUU-AAAUGAGACUC----------------ACAUUC-CAAA-CAUUUUCAAUUUGAAAUC----- ....((.((((((((((..........((((((((...))))))))........-)))))))....----------------.....)-)).)-)..((((.....))))..----- ( -12.87) >DroEre_CAF1 14388 108 + 1 CA-GGACAUGAUCCUAUUAUGGAAAGAAACAAAUGACUCAUUUGUUUCAAAAUUUCGGC------CAAACAAUGAAUUGUC-ACAUUA-CGUAUCUUUUCCAAAUUGAAAUGAUACA ..-.((((...(((......)))..((((((((((...))))))))))...........------............))))-......-.(((((.((((......)))).))))). ( -24.20) >DroYak_CAF1 14460 110 + 1 AU-CAAAACAAUCAUUGUAUGCAAAUAAACAAAUGACUCAUUUGUUUCAGAAUUUCGGCAUGAUUUAAAAAAACAAUUGCUGAGAC------UCAUUUUCUAAUUUGAAAUAACACA .(-((((...................(((((((((...)))))))))..((.((((((((.................)))))))).------)).........)))))......... ( -18.03) >consensus CA_AGUCUGGCUCAUUUUAUGCAAAUAAACAAAUGACUCAUUUGUUUCAAAAUU_CGAUGAGACUCAA__AA_____UG___ACAUUC_CAAA_AAUUUUCAAUUUGAAAUC_____ ..........(((((((.........(((((((((...)))))))))........)))))))....................................................... ( -5.19 = -7.23 + 2.04)


| Location | 5,524,343 – 5,524,439 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -12.42 |
| Consensus MFE | -8.32 |
| Energy contribution | -8.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5524343 96 + 22224390 UCAUUUGUUUAAAAAUUCGAUGAGACUCAAGGAAUGUCACAUUCACAAAAAUUUUCAAUUUGAAAUCUGAAAAGAUUCAACAGCAUAAGCAAAUGA (((((((((((.........(((((......(((((...)))))........)))))..(((((.(((....)))))))).....))))))))))) ( -20.24) >DroSec_CAF1 14217 86 + 1 UCAUUUGUUCCAAAUUUAAAUGAGACUC----------ACAUUCACAAAUAUUUUCAAUUUGAAAUCUAAAAUGAUUCAACAGCAUCCACAAAUGA ((((((((............(((((...----------..............)))))..(((((.((......)))))))........)))))))) ( -9.43) >DroSim_CAF1 16756 85 + 1 UCAUUUGUUCCAAAUUUAAAUGAGACUC----------ACAUUC-CAAACAUUUUCAAUUUGAAAUCUAAAAUGAUUCAACAGCAUCCGCAUAUGA (((((((.........))))))).....----------......-.........(((..(((((.((......)))))))..((....))...))) ( -7.60) >consensus UCAUUUGUUCCAAAUUUAAAUGAGACUC__________ACAUUCACAAAAAUUUUCAAUUUGAAAUCUAAAAUGAUUCAACAGCAUCCGCAAAUGA ((((((((............(((((...........................)))))..(((((.((......)))))))........)))))))) ( -8.32 = -8.43 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:12 2006