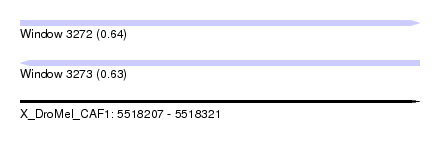
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,518,207 – 5,518,321 |
| Length | 114 |
| Max. P | 0.636409 |

| Location | 5,518,207 – 5,518,321 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -19.21 |
| Energy contribution | -21.10 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
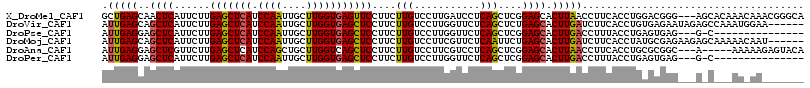
>X_DroMel_CAF1 5518207 114 + 22224390 UGCCCGUUUGUUUGUGCU---CCCGUCCAGGUGAAGGUUAAGUGCUCCGAGCUGAGGAUCAAGGACAAGAAGGAACUCACCAAGCAAUUGGAUGAGCUCAAGAAUGAGUUGCUCAGC .....((((.(((.((.(---((.((((.(((...((.........))..)))..))))...))))).))).))))...((((....)))).((((((((....)))...))))).. ( -28.10) >DroVir_CAF1 9854 111 + 1 ------UUCCAUUUGGCUCUAUUCUCACAGGUGAAGAUCAAGUGCUCAGAGCUGAGAACCAAGGACAAGAAGGAGCUCACCAAGCAAUUGGAUGAGCUCAAGAAUGAGCUGCUCAAU ------........(((((.(((((....(((............((((....)))).)))............(((((((((((....)))).))))))).))))))))))....... ( -33.72) >DroPse_CAF1 23089 98 + 1 ---------------G-C---CUCACUCAGGUAAAGGUCAAGUGCUCCGAGCUGAGAACCAAGGACAAGAAGGAGCUCACCAAGCAAUUGGAUGAGCUCAAGAAUGAGCUCCUCAAU ---------------(-(---.((((((((.(...((.........)).).)))))................(((((((((((....)))).))))))).....)))))........ ( -29.50) >DroMoj_CAF1 8669 111 + 1 ------AUUGUUUUUGCUCUUCUCGCAUAGGUGAAGAUCAAGUGCUCAGAAUUGAGAACGAAGGACAAGAAGGAGCUCACCAAGCAAUUGGAUGAGCUCAAGAAUGAGCUGCUCAAU ------....((((((..((((((((....)))).......((.(((......))).))))))..)))))).(((((((((((....)))).))))))).....((((...)))).. ( -35.90) >DroAna_CAF1 7419 109 + 1 UGUACUCUUUUU-----U---GCCGCGCAGGUGAAGGUUAAGUGCUCCGAGCUGAGGACGAAGGACAAGAAGGAGCUGACCAAGCAGCUGGAUGAGCUCAAGAACGAGCUCCUCAAU .....(((((((-----(---(((((((.............))))(((.......)))....)).)))))))))((((......))))..((.((((((......)))))).))... ( -33.52) >DroPer_CAF1 25604 98 + 1 ---------------G-C---CUCACUCAGGUAAAGGUCAAGUGCUCCGAGCUGAGAACCAAGGACAAGAAGGAGCUCACCAAGCAAUUGGAUGAGCUCAAGAAUGAGCUCCUCAAU ---------------(-(---.((((((((.(...((.........)).).)))))................(((((((((((....)))).))))))).....)))))........ ( -29.50) >consensus _______UU_UU___G_U___CUCGCACAGGUGAAGGUCAAGUGCUCCGAGCUGAGAACCAAGGACAAGAAGGAGCUCACCAAGCAAUUGGAUGAGCUCAAGAAUGAGCUCCUCAAU ....................................(((..((.(((......))).))....)))......(((((((((((....)))).))))))).....((((...)))).. (-19.21 = -21.10 + 1.89)

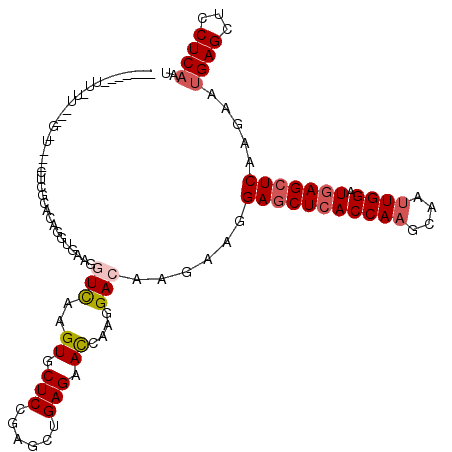

| Location | 5,518,207 – 5,518,321 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.85 |
| Mean single sequence MFE | -30.39 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.42 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5518207 114 - 22224390 GCUGAGCAACUCAUUCUUGAGCUCAUCCAAUUGCUUGGUGAGUUCCUUCUUGUCCUUGAUCCUCAGCUCGGAGCACUUAACCUUCACCUGGACGGG---AGCACAAACAAACGGGCA ((((((...)))......(((((((.((((....))))))))))).((((((((((((.....)))...((((........))))....)))))))---))............))). ( -28.50) >DroVir_CAF1 9854 111 - 1 AUUGAGCAGCUCAUUCUUGAGCUCAUCCAAUUGCUUGGUGAGCUCCUUCUUGUCCUUGGUUCUCAGCUCUGAGCACUUGAUCUUCACCUGUGAGAAUAGAGCCAAAUGGAA------ ...((((.(((((....)))))(((.((((....))))))))))).......(((((((((((...(((...(((..((.....))..))))))...))))))))..))).------ ( -34.20) >DroPse_CAF1 23089 98 - 1 AUUGAGGAGCUCAUUCUUGAGCUCAUCCAAUUGCUUGGUGAGCUCCUUCUUGUCCUUGGUUCUCAGCUCGGAGCACUUGACCUUUACCUGAGUGAG---G-C--------------- ...((((((((((((...((((..........))))))))))))))))...(((....(((((......)))))....)))((((((....)))))---)-.--------------- ( -30.80) >DroMoj_CAF1 8669 111 - 1 AUUGAGCAGCUCAUUCUUGAGCUCAUCCAAUUGCUUGGUGAGCUCCUUCUUGUCCUUCGUUCUCAAUUCUGAGCACUUGAUCUUCACCUAUGCGAGAAGAGCAAAAACAAU------ ........((((......(((((((.((((....))))))))))).(((((((.....((.((((....)))).)).((.....)).....))))))))))).........------ ( -30.90) >DroAna_CAF1 7419 109 - 1 AUUGAGGAGCUCGUUCUUGAGCUCAUCCAGCUGCUUGGUCAGCUCCUUCUUGUCCUUCGUCCUCAGCUCGGAGCACUUAACCUUCACCUGCGCGGC---A-----AAAAAGAGUACA ...((.(((((((....))))))).))..(((((..(((..(((((...(((...........)))...)))))...........)))...)))))---.-----............ ( -27.12) >DroPer_CAF1 25604 98 - 1 AUUGAGGAGCUCAUUCUUGAGCUCAUCCAAUUGCUUGGUGAGCUCCUUCUUGUCCUUGGUUCUCAGCUCGGAGCACUUGACCUUUACCUGAGUGAG---G-C--------------- ...((((((((((((...((((..........))))))))))))))))...(((....(((((......)))))....)))((((((....)))))---)-.--------------- ( -30.80) >consensus AUUGAGCAGCUCAUUCUUGAGCUCAUCCAAUUGCUUGGUGAGCUCCUUCUUGUCCUUGGUUCUCAGCUCGGAGCACUUGACCUUCACCUGAGCGAG___A_C___AA_AA_______ .(((((..((((......(((((((.((((....)))))))))))....(((...........)))....)))).)))))..................................... (-18.55 = -18.42 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:07 2006